| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002192_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_048635_010 |
0.0021871817 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_081212_010 |
0.0334536542 |
- |
- |
- |
| 4 |
Hb_000742_050 |
0.0381009791 |
- |
- |
hypothetical protein CL1373Contig1_03, partial [Pinus lambertiana] |
| 5 |
Hb_049719_010 |
0.0407584793 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_039385_010 |
0.0414408119 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_011903_030 |
0.0436956728 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Populus euphratica] |
| 8 |
Hb_002539_060 |
0.0447996868 |
- |
- |
- |
| 9 |
Hb_000629_050 |
0.0456548338 |
- |
- |
- |
| 10 |
Hb_003687_260 |
0.045765661 |
- |
- |
PREDICTED: inorganic phosphate transporter 1-4-like [Camelina sativa] |
| 11 |
Hb_005417_020 |
0.0463744682 |
- |
- |
- |
| 12 |
Hb_129319_010 |
0.0465629597 |
- |
- |
PREDICTED: uncharacterized protein LOC104115096 [Nicotiana tomentosiformis] |
| 13 |
Hb_005171_030 |
0.0467687633 |
- |
- |
hypothetical protein EUGRSUZ_H02434 [Eucalyptus grandis] |
| 14 |
Hb_025248_030 |
0.056453672 |
- |
- |
hypothetical protein JCGZ_09377 [Jatropha curcas] |
| 15 |
Hb_173479_010 |
0.0580586097 |
- |
- |
- |
| 16 |
Hb_002232_330 |
0.0581174397 |
- |
- |
- |
| 17 |
Hb_000522_100 |
0.0583258925 |
- |
- |
PREDICTED: putative SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3-like 3 [Nelumbo nucifera] |
| 18 |
Hb_001198_070 |
0.062727157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004712_100 |
0.067900028 |
- |
- |
hypothetical protein CICLE_v10003434mg, partial [Citrus clementina] |
| 20 |
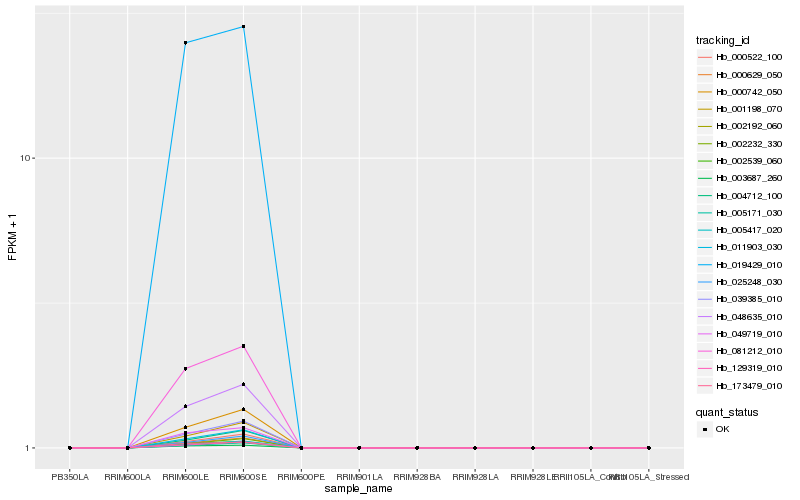
Hb_019429_010 |
0.0790727663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |