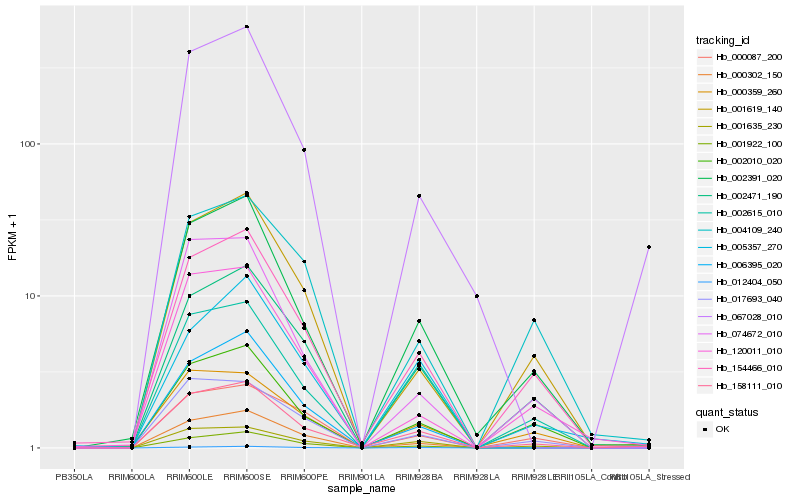
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158111_010 |
0.0 |
- |
- |
S-locus-specific glycoprotein precursor, putative [Ricinus communis] |
| 2 |
Hb_154466_010 |
0.1004388435 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 3 |
Hb_002391_020 |
0.1030890512 |
transcription factor |
TF Family: NAC |
PREDICTED: putative NAC domain-containing protein 94 [Jatropha curcas] |
| 4 |
Hb_002471_190 |
0.1123607392 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 5 |
Hb_001635_230 |
0.1137337555 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_002615_010 |
0.1198098129 |
- |
- |
Uncharacterized protein TCM_040942 [Theobroma cacao] |
| 7 |
Hb_002010_020 |
0.1261820811 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 8 |
Hb_017693_040 |
0.1275370606 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 9 |
Hb_000302_150 |
0.1334192955 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1581940 [Ricinus communis] |
| 10 |
Hb_001619_140 |
0.1360604502 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 11 |
Hb_006395_020 |
0.1398388649 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 12 |
Hb_000359_260 |
0.1444660803 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 13 |
Hb_001922_100 |
0.1445961044 |
- |
- |
S-type anion channel SLAH2 [Triticum urartu] |
| 14 |
Hb_067028_010 |
0.1468379379 |
- |
- |
BnaC03g72040D [Brassica napus] |
| 15 |
Hb_005357_270 |
0.1529969069 |
- |
- |
Glucose-6-phosphate/phosphate translocator 2, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_120011_010 |
0.1531648189 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 17 |
Hb_012404_050 |
0.1537899444 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 18 |
Hb_004109_240 |
0.1542021489 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 19 |
Hb_000087_200 |
0.1547653287 |
- |
- |
PREDICTED: uncharacterized protein LOC105641954 [Jatropha curcas] |
| 20 |
Hb_074672_010 |
0.1550712578 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |