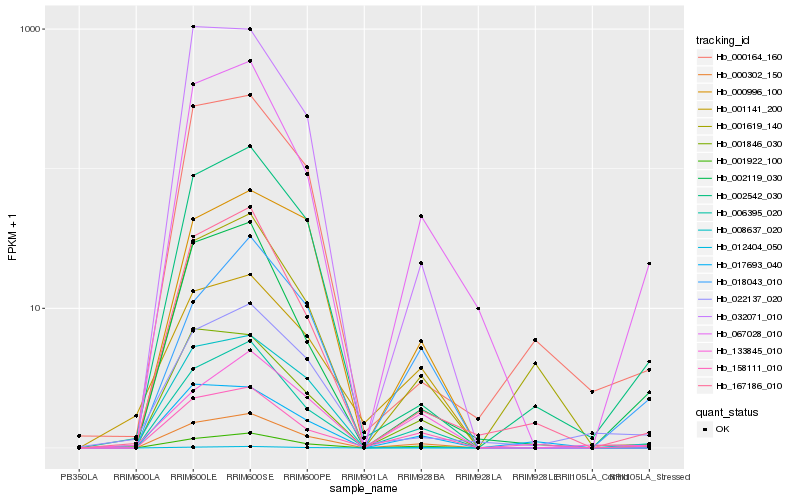
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000302_150 |
0.0 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1581940 [Ricinus communis] |
| 2 |
Hb_001922_100 |
0.048210123 |
- |
- |
S-type anion channel SLAH2 [Triticum urartu] |
| 3 |
Hb_067028_010 |
0.0988068174 |
- |
- |
BnaC03g72040D [Brassica napus] |
| 4 |
Hb_006395_020 |
0.1150438759 |
- |
- |
hypothetical protein RCOM_0725510 [Ricinus communis] |
| 5 |
Hb_158111_010 |
0.1334192955 |
- |
- |
S-locus-specific glycoprotein precursor, putative [Ricinus communis] |
| 6 |
Hb_002542_030 |
0.1374579517 |
transcription factor |
TF Family: Tify |
JAZ7 [Hevea brasiliensis] |
| 7 |
Hb_002119_030 |
0.1429700722 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 8 |
Hb_001846_030 |
0.1434899197 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 9 |
Hb_018043_010 |
0.1467513549 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 10 |
Hb_012404_050 |
0.1559336459 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 11 |
Hb_167186_010 |
0.1570484284 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 12 |
Hb_000164_160 |
0.1662683581 |
transcription factor |
TF Family: GRAS |
PREDICTED: chitin-inducible gibberellin-responsive protein 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001619_140 |
0.1665936645 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 14 |
Hb_022137_020 |
0.167335153 |
transcription factor |
TF Family: AP2 |
PREDICTED: dehydration-responsive element-binding protein 1B-like [Jatropha curcas] |
| 15 |
Hb_133845_010 |
0.1688330599 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_001141_200 |
0.1690542316 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: parafibromin [Prunus mume] |
| 17 |
Hb_032071_010 |
0.1700631242 |
- |
- |
hypothetical protein EUGRSUZ_J02969 [Eucalyptus grandis] |
| 18 |
Hb_017693_040 |
0.1720387321 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 19 |
Hb_000996_100 |
0.1720525586 |
- |
- |
Nucleotide binding, putative [Theobroma cacao] |
| 20 |
Hb_008637_020 |
0.1731149133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |