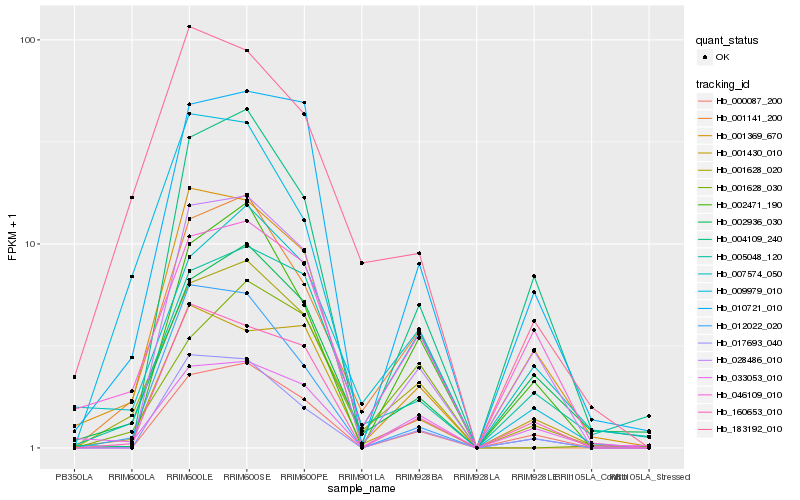
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001628_020 |
0.0 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 2 |
Hb_001141_200 |
0.1141697056 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: parafibromin [Prunus mume] |
| 3 |
Hb_010721_010 |
0.1451112831 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Populus euphratica] |
| 4 |
Hb_002936_030 |
0.1506844035 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007574_050 |
0.1522289293 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 6 |
Hb_000087_200 |
0.1522455302 |
- |
- |
PREDICTED: uncharacterized protein LOC105641954 [Jatropha curcas] |
| 7 |
Hb_005048_120 |
0.1523927925 |
- |
- |
- |
| 8 |
Hb_183192_010 |
0.1595385969 |
- |
- |
PREDICTED: thioredoxin reductase NTRC [Sesamum indicum] |
| 9 |
Hb_028486_010 |
0.1622594001 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 10 |
Hb_001369_670 |
0.1623352968 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 11 |
Hb_046109_010 |
0.1642997643 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 12 |
Hb_002471_190 |
0.1665755199 |
- |
- |
PREDICTED: U-box domain-containing protein 21-like [Jatropha curcas] |
| 13 |
Hb_160653_010 |
0.1673432387 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 14 |
Hb_009979_010 |
0.1677543261 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 15 |
Hb_033053_010 |
0.1722067172 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 16 |
Hb_004109_240 |
0.1722239103 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 17 |
Hb_001628_030 |
0.1739364554 |
- |
- |
PREDICTED: uncharacterized protein LOC105629266 isoform X1 [Jatropha curcas] |
| 18 |
Hb_012022_020 |
0.1758413309 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0006s10190g [Populus trichocarpa] |
| 19 |
Hb_001430_010 |
0.1772916182 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_017693_040 |
0.1773485022 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |