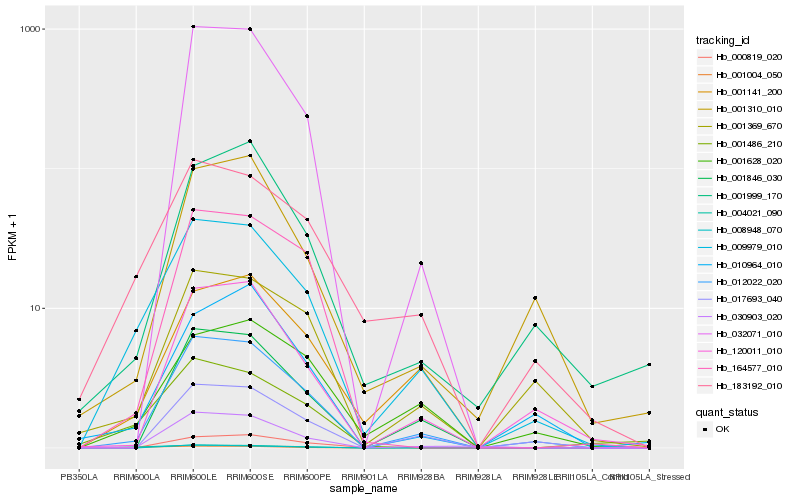
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009979_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 2 |
Hb_183192_010 |
0.1209805738 |
- |
- |
PREDICTED: thioredoxin reductase NTRC [Sesamum indicum] |
| 3 |
Hb_012022_020 |
0.1261343517 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0006s10190g [Populus trichocarpa] |
| 4 |
Hb_004021_090 |
0.1674395582 |
- |
- |
hypothetical protein JCGZ_09233 [Jatropha curcas] |
| 5 |
Hb_001628_020 |
0.1677543261 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 6 |
Hb_001141_200 |
0.1680969946 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: parafibromin [Prunus mume] |
| 7 |
Hb_001004_050 |
0.171146972 |
- |
- |
PREDICTED: uncharacterized protein LOC103335231 [Prunus mume] |
| 8 |
Hb_001369_670 |
0.1795260525 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 9 |
Hb_000819_020 |
0.1815784073 |
- |
- |
hypothetical protein PRUPE_ppa020290mg, partial [Prunus persica] |
| 10 |
Hb_001486_210 |
0.1844083724 |
- |
- |
- |
| 11 |
Hb_001846_030 |
0.1889220664 |
- |
- |
PREDICTED: cytochrome P450 94A2-like [Jatropha curcas] |
| 12 |
Hb_017693_040 |
0.1944960709 |
- |
- |
tyrosine kinase family protein [Medicago truncatula] |
| 13 |
Hb_001999_170 |
0.1972844536 |
- |
- |
PREDICTED: uncharacterized protein LOC105630609 [Jatropha curcas] |
| 14 |
Hb_030903_020 |
0.1974804031 |
- |
- |
PREDICTED: putative 12-oxophytodienoate reductase 11 [Jatropha curcas] |
| 15 |
Hb_032071_010 |
0.1975492232 |
- |
- |
hypothetical protein EUGRSUZ_J02969 [Eucalyptus grandis] |
| 16 |
Hb_120011_010 |
0.197843261 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 17 |
Hb_001310_010 |
0.2011882318 |
- |
- |
PREDICTED: calcium-transporting ATPase 2, plasma membrane-type [Jatropha curcas] |
| 18 |
Hb_008948_070 |
0.2021682619 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_164577_010 |
0.2026605884 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 20 |
Hb_010964_010 |
0.204179326 |
- |
- |
- |