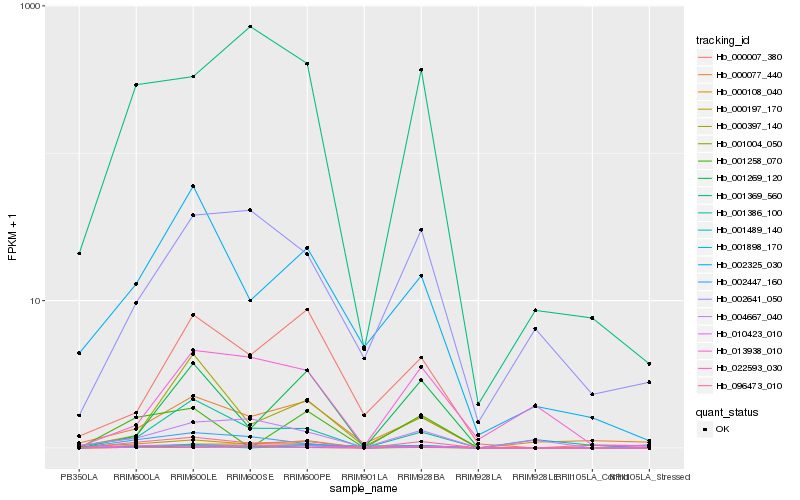
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_096473_010 |
0.0 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 2 |
Hb_010423_010 |
0.1620217887 |
- |
- |
- |
| 3 |
Hb_000108_040 |
0.2286143042 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Gossypium raimondii] |
| 4 |
Hb_000197_170 |
0.2360687149 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_001004_050 |
0.2389087483 |
- |
- |
PREDICTED: uncharacterized protein LOC103335231 [Prunus mume] |
| 6 |
Hb_022593_030 |
0.250407417 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 7 |
Hb_001369_560 |
0.2507058815 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 8 |
Hb_002325_030 |
0.2614459582 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 2 [Jatropha curcas] |
| 9 |
Hb_000077_440 |
0.2638923656 |
- |
- |
PREDICTED: ELMO domain-containing protein A isoform X1 [Jatropha curcas] |
| 10 |
Hb_001269_120 |
0.2704859532 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF003-like [Jatropha curcas] |
| 11 |
Hb_001489_140 |
0.2721157367 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_21694 [Jatropha curcas] |
| 12 |
Hb_001386_100 |
0.2733435488 |
transcription factor |
TF Family: M-type |
PREDICTED: MADS-box transcription factor PHERES 1-like [Jatropha curcas] |
| 13 |
Hb_000007_380 |
0.2754963543 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 14 |
Hb_000397_140 |
0.2772203872 |
transcription factor |
TF Family: PLATZ |
hypothetical protein JCGZ_16980 [Jatropha curcas] |
| 15 |
Hb_001258_070 |
0.2808037809 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002641_050 |
0.2900994867 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 48 [Jatropha curcas] |
| 17 |
Hb_004667_040 |
0.2922419531 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_002447_160 |
0.2943428994 |
- |
- |
- |
| 19 |
Hb_001898_170 |
0.2989206553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_013938_010 |
0.3030245127 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like isoform X3 [Citrus sinensis] |