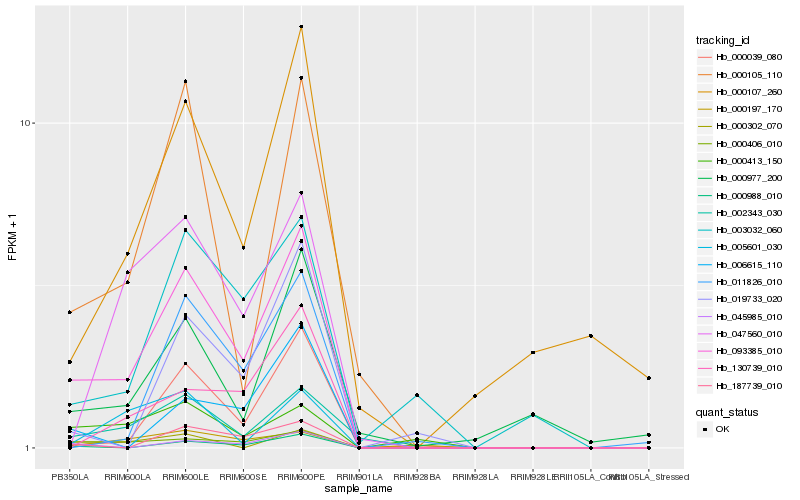
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_093385_010 |
0.0 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Pyrus x bretschneideri] |
| 2 |
Hb_000406_010 |
0.1463857938 |
- |
- |
PREDICTED: uncharacterized protein LOC105647846 [Jatropha curcas] |
| 3 |
Hb_002343_030 |
0.1887974262 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000988_010 |
0.2048810055 |
- |
- |
hypothetical protein POPTR_0004s17730g [Populus trichocarpa] |
| 5 |
Hb_000039_080 |
0.207920403 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 6 |
Hb_000105_110 |
0.2097466617 |
- |
- |
hypothetical protein, partial [Fervidibacteria bacterium SCGC AAA471-D06] |
| 7 |
Hb_187739_010 |
0.2127137862 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 8 |
Hb_045985_010 |
0.224554346 |
- |
- |
hypothetical protein JCGZ_00291 [Jatropha curcas] |
| 9 |
Hb_003032_060 |
0.2263834493 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 10 |
Hb_000413_150 |
0.2358008631 |
- |
- |
PREDICTED: uncharacterized protein LOC103709996 [Phoenix dactylifera] |
| 11 |
Hb_000197_170 |
0.2378944398 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000107_260 |
0.2403501297 |
- |
- |
- |
| 13 |
Hb_006615_110 |
0.2433446 |
transcription factor |
TF Family: bZIP |
hypothetical protein JCGZ_11777 [Jatropha curcas] |
| 14 |
Hb_047560_010 |
0.2434752861 |
- |
- |
PREDICTED: GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 15 |
Hb_130739_010 |
0.248242966 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 16 |
Hb_019733_020 |
0.2517610129 |
- |
- |
hypothetical protein POPTR_0003s15470g [Populus trichocarpa] |
| 17 |
Hb_000302_070 |
0.2518369233 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 18 |
Hb_011826_010 |
0.2566820036 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 19 |
Hb_000977_200 |
0.2570135862 |
- |
- |
PREDICTED: acyl-[acyl-carrier-protein] desaturase, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_005601_030 |
0.2577366045 |
- |
- |
organic anion transporter, putative [Ricinus communis] |