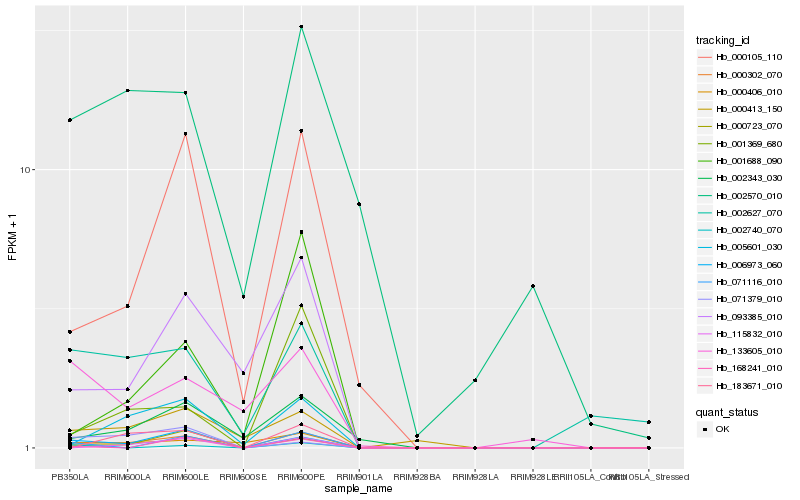
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000302_070 |
0.0 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 2 |
Hb_006973_060 |
0.1646023959 |
- |
- |
Uncharacterized protein TCM_038475 [Theobroma cacao] |
| 3 |
Hb_000723_070 |
0.1944883119 |
- |
- |
PREDICTED: thylakoid lumenal 15.0 kDa protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_000105_110 |
0.2056744164 |
- |
- |
hypothetical protein, partial [Fervidibacteria bacterium SCGC AAA471-D06] |
| 5 |
Hb_005601_030 |
0.2358754712 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 6 |
Hb_093385_010 |
0.2518369233 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Pyrus x bretschneideri] |
| 7 |
Hb_002343_030 |
0.2613856318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000413_150 |
0.2631303716 |
- |
- |
PREDICTED: uncharacterized protein LOC103709996 [Phoenix dactylifera] |
| 9 |
Hb_071116_010 |
0.2635055365 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 10 |
Hb_115832_010 |
0.2674072801 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000406_010 |
0.2739730064 |
- |
- |
PREDICTED: uncharacterized protein LOC105647846 [Jatropha curcas] |
| 12 |
Hb_002627_070 |
0.279280521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001369_680 |
0.279447208 |
- |
- |
PREDICTED: uncharacterized protein LOC105648582 [Jatropha curcas] |
| 14 |
Hb_183671_010 |
0.2854011625 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 15 |
Hb_002570_010 |
0.2965107485 |
- |
- |
hypothetical protein JCGZ_25329 [Jatropha curcas] |
| 16 |
Hb_133605_010 |
0.2968949854 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 17 |
Hb_001688_090 |
0.2980745962 |
- |
- |
alcohol dehydrogenase 7 [Vitis vinifera] |
| 18 |
Hb_168241_010 |
0.3012876371 |
- |
- |
hypothetical protein POPTR_0013s14620g, partial [Populus trichocarpa] |
| 19 |
Hb_071379_010 |
0.3062326644 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 3-like [Nelumbo nucifera] |
| 20 |
Hb_002740_070 |
0.3063896429 |
- |
- |
nitrate transporter, putative [Ricinus communis] |