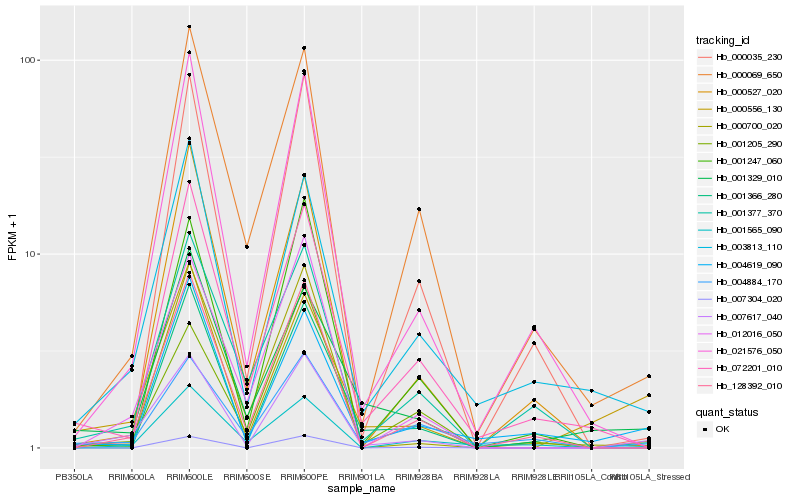
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007617_040 |
0.0 |
- |
- |
PREDICTED: syntaxin-112-like [Jatropha curcas] |
| 2 |
Hb_012016_050 |
0.1687161086 |
- |
- |
hypothetical protein JCGZ_09036 [Jatropha curcas] |
| 3 |
Hb_000556_130 |
0.174012557 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |
| 4 |
Hb_128392_010 |
0.1755415832 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 5 |
Hb_021576_050 |
0.1769091198 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 6 |
Hb_003813_110 |
0.1850027717 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001247_060 |
0.191798514 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_004619_090 |
0.1927886352 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001366_280 |
0.1928261189 |
- |
- |
PREDICTED: uncharacterized protein LOC105642732 [Jatropha curcas] |
| 10 |
Hb_000700_020 |
0.193622085 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 11 |
Hb_007304_020 |
0.1946881586 |
- |
- |
PREDICTED: uncharacterized protein LOC100256879 [Vitis vinifera] |
| 12 |
Hb_000035_230 |
0.1950469313 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 13 |
Hb_004884_170 |
0.1975686942 |
- |
- |
hypothetical protein AALP_AA1G067000 [Arabis alpina] |
| 14 |
Hb_000069_650 |
0.197863425 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000527_020 |
0.2003626612 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 16 |
Hb_001377_370 |
0.2009659077 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 17 |
Hb_001329_010 |
0.203116417 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_001205_290 |
0.2032820314 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 19 |
Hb_072201_010 |
0.2055133439 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 20 |
Hb_001565_090 |
0.2056039283 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |