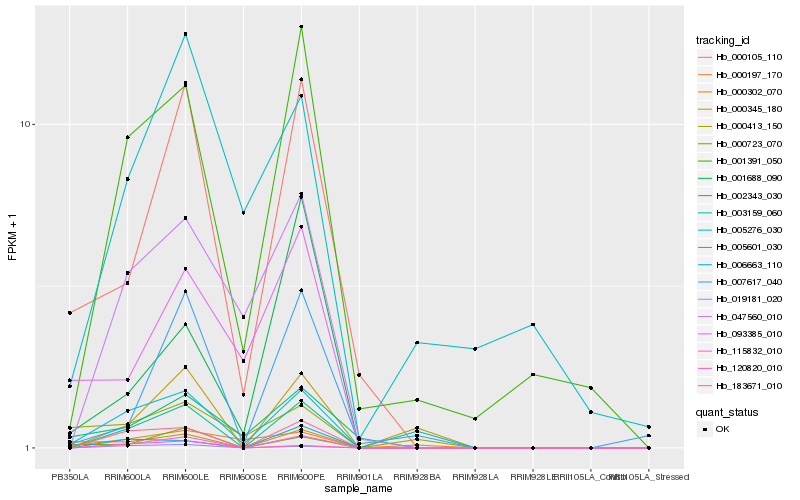
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005601_030 |
0.0 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 2 |
Hb_183671_010 |
0.1520309884 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 3 |
Hb_115832_010 |
0.1726267362 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_001391_050 |
0.1903663488 |
- |
- |
PREDICTED: probable carboxylesterase 18 [Jatropha curcas] |
| 5 |
Hb_019181_020 |
0.2025834483 |
- |
- |
PREDICTED: uncharacterized protein LOC105763068 [Gossypium raimondii] |
| 6 |
Hb_047560_010 |
0.2038854764 |
- |
- |
PREDICTED: GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 7 |
Hb_000105_110 |
0.2179275148 |
- |
- |
hypothetical protein, partial [Fervidibacteria bacterium SCGC AAA471-D06] |
| 8 |
Hb_002343_030 |
0.2338424414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000302_070 |
0.2358754712 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 10 |
Hb_120820_010 |
0.2391655566 |
- |
- |
PREDICTED: uncharacterized protein LOC105785094 [Gossypium raimondii] |
| 11 |
Hb_006663_110 |
0.2432939901 |
- |
- |
PREDICTED: U-box domain-containing protein 34-like [Camelina sativa] |
| 12 |
Hb_000723_070 |
0.2509628194 |
- |
- |
PREDICTED: thylakoid lumenal 15.0 kDa protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_093385_010 |
0.2577366045 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Pyrus x bretschneideri] |
| 14 |
Hb_000197_170 |
0.2728413495 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000413_150 |
0.2813597594 |
- |
- |
PREDICTED: uncharacterized protein LOC103709996 [Phoenix dactylifera] |
| 16 |
Hb_000345_180 |
0.2843930582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005276_030 |
0.2905949531 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 18 |
Hb_001688_090 |
0.2939214498 |
- |
- |
alcohol dehydrogenase 7 [Vitis vinifera] |
| 19 |
Hb_003159_060 |
0.294385914 |
- |
- |
Uncharacterized protein TCM_001961 [Theobroma cacao] |
| 20 |
Hb_007617_040 |
0.297407887 |
- |
- |
PREDICTED: syntaxin-112-like [Jatropha curcas] |