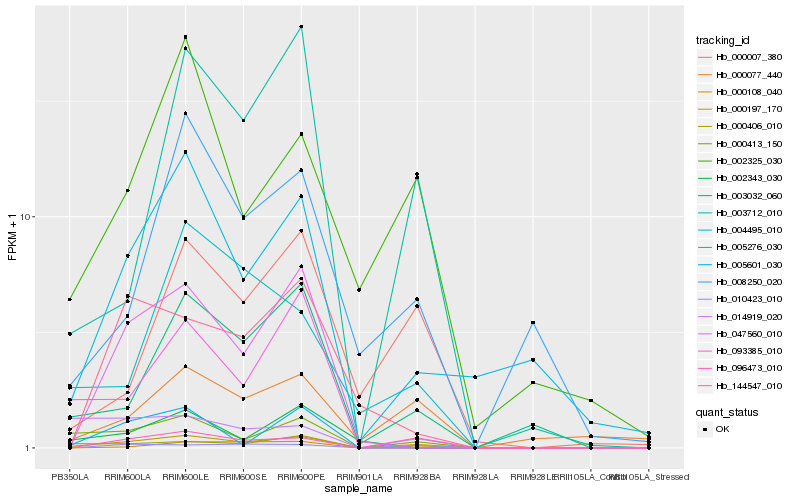
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000197_170 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_000413_150 |
0.1731723048 |
- |
- |
PREDICTED: uncharacterized protein LOC103709996 [Phoenix dactylifera] |
| 3 |
Hb_003032_060 |
0.2049799635 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 4 |
Hb_005276_030 |
0.2215040258 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 5 |
Hb_047560_010 |
0.2274522207 |
- |
- |
PREDICTED: GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 6 |
Hb_096473_010 |
0.2360687149 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 7 |
Hb_093385_010 |
0.2378944398 |
- |
- |
PREDICTED: probable mannitol dehydrogenase [Pyrus x bretschneideri] |
| 8 |
Hb_003712_010 |
0.2407840114 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_000406_010 |
0.2525014258 |
- |
- |
PREDICTED: uncharacterized protein LOC105647846 [Jatropha curcas] |
| 10 |
Hb_002325_030 |
0.2580826804 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 2 [Jatropha curcas] |
| 11 |
Hb_144547_010 |
0.2625719564 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 12 |
Hb_008250_020 |
0.2639371296 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_004495_010 |
0.2653971186 |
- |
- |
PREDICTED: thioredoxin reductase NTRC [Sesamum indicum] |
| 14 |
Hb_010423_010 |
0.2683295491 |
- |
- |
- |
| 15 |
Hb_000007_380 |
0.2687165332 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 16 |
Hb_000077_440 |
0.2697152559 |
- |
- |
PREDICTED: ELMO domain-containing protein A isoform X1 [Jatropha curcas] |
| 17 |
Hb_002343_030 |
0.2704499142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005601_030 |
0.2728413495 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 19 |
Hb_014919_020 |
0.2740315654 |
- |
- |
- |
| 20 |
Hb_000108_040 |
0.2753852154 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Gossypium raimondii] |