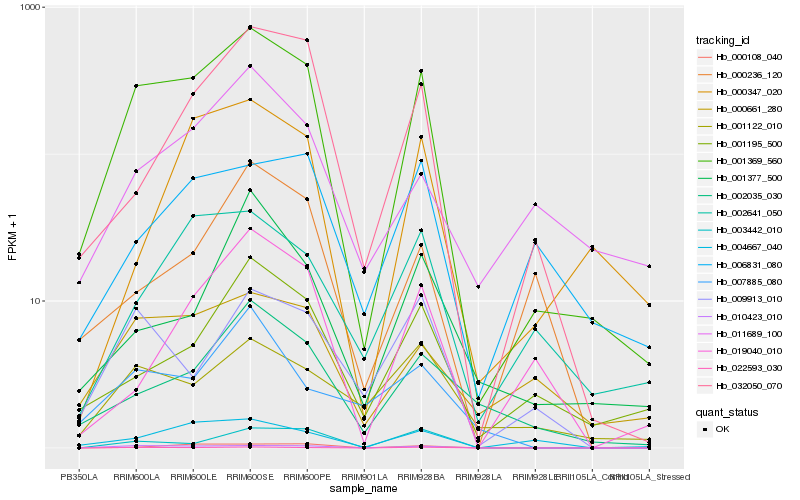
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_560 |
0.0 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 2 |
Hb_022593_030 |
0.1447601404 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 3 |
Hb_010423_010 |
0.1616602855 |
- |
- |
- |
| 4 |
Hb_032050_070 |
0.2110718675 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 5 |
Hb_002035_030 |
0.214471931 |
- |
- |
hypothetical protein PRUPE_ppa003221mg [Prunus persica] |
| 6 |
Hb_003442_010 |
0.2164721855 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 4-like isoform X2 [Citrus sinensis] |
| 7 |
Hb_000661_280 |
0.2165421343 |
- |
- |
PREDICTED: protein TRAUCO [Jatropha curcas] |
| 8 |
Hb_004667_040 |
0.2167219505 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_001195_500 |
0.2179865986 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 10 |
Hb_000108_040 |
0.2197126692 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Gossypium raimondii] |
| 11 |
Hb_009913_010 |
0.2235885005 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_002641_050 |
0.2253358404 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 48 [Jatropha curcas] |
| 13 |
Hb_001122_010 |
0.2316913866 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 14 |
Hb_000347_020 |
0.2363478826 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 15 |
Hb_019040_010 |
0.2375799639 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG7 [Jatropha curcas] |
| 16 |
Hb_001377_500 |
0.2377344199 |
- |
- |
DOF AFFECTING GERMINATION 2 family protein [Populus trichocarpa] |
| 17 |
Hb_011689_100 |
0.2438620145 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 18 |
Hb_007885_080 |
0.2444154367 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006831_080 |
0.2455414883 |
- |
- |
PREDICTED: uncharacterized protein LOC105642126 [Jatropha curcas] |
| 20 |
Hb_000236_120 |
0.2458983686 |
- |
- |
- |