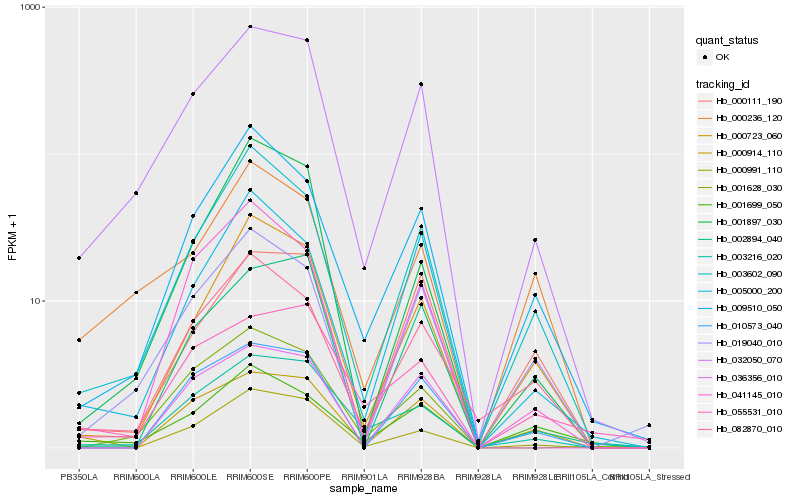
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032050_070 |
0.0 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 2 |
Hb_003216_020 |
0.1320101966 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 3 |
Hb_019040_010 |
0.1354445298 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG7 [Jatropha curcas] |
| 4 |
Hb_003602_090 |
0.1355589987 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 5 |
Hb_001628_030 |
0.1411762652 |
- |
- |
PREDICTED: uncharacterized protein LOC105629266 isoform X1 [Jatropha curcas] |
| 6 |
Hb_009510_050 |
0.1519510477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000723_060 |
0.1585268018 |
- |
- |
PREDICTED: uncharacterized protein LOC105642243 [Jatropha curcas] |
| 8 |
Hb_082870_010 |
0.1586882491 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 9 |
Hb_005000_200 |
0.1610938263 |
- |
- |
PREDICTED: uncharacterized protein LOC105637887 [Jatropha curcas] |
| 10 |
Hb_000991_110 |
0.162412767 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 11 |
Hb_000914_110 |
0.1643600346 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2 [Jatropha curcas] |
| 12 |
Hb_010573_040 |
0.1646418025 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal isoform X2 [Populus euphratica] |
| 13 |
Hb_002894_040 |
0.1662004883 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_041145_010 |
0.1668924057 |
- |
- |
hypothetical protein CISIN_1g021653mg [Citrus sinensis] |
| 15 |
Hb_000236_120 |
0.1670637707 |
- |
- |
- |
| 16 |
Hb_000111_190 |
0.171175334 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1 [Jatropha curcas] |
| 17 |
Hb_001897_030 |
0.1717189826 |
- |
- |
PREDICTED: actin-depolymerizing factor 5 [Jatropha curcas] |
| 18 |
Hb_055531_010 |
0.1718732359 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Pyrus x bretschneideri] |
| 19 |
Hb_001699_050 |
0.1747046468 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s11410g [Populus trichocarpa] |
| 20 |
Hb_036356_010 |
0.1750053314 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |