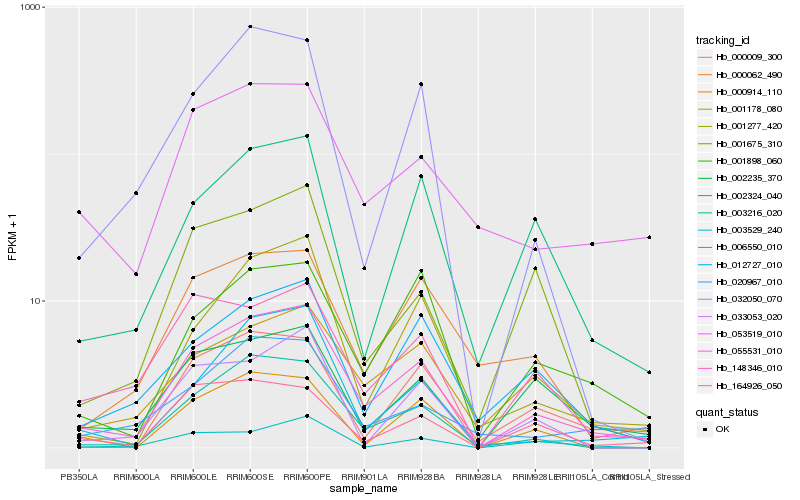
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_055531_010 |
0.0 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Pyrus x bretschneideri] |
| 2 |
Hb_003216_020 |
0.1317501132 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 3 |
Hb_001277_420 |
0.1595481167 |
- |
- |
PREDICTED: protein lin-12-like isoform X2 [Populus euphratica] |
| 4 |
Hb_002235_370 |
0.161387888 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 5 |
Hb_006550_010 |
0.1619162197 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Eucalyptus grandis] |
| 6 |
Hb_012727_010 |
0.1652644722 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 7 |
Hb_148346_010 |
0.1664769684 |
- |
- |
- |
| 8 |
Hb_001178_080 |
0.1673127784 |
- |
- |
PREDICTED: protein kinase PVPK-1 [Jatropha curcas] |
| 9 |
Hb_001898_060 |
0.168252506 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002324_040 |
0.1697440602 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 11 |
Hb_032050_070 |
0.1718732359 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 12 |
Hb_164926_050 |
0.1721753971 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 13 |
Hb_053519_010 |
0.1724893265 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |
| 14 |
Hb_003529_240 |
0.1725165261 |
transcription factor |
TF Family: AP2 |
PREDICTED: ethylene-responsive transcription factor RAP2-7-like isoform X2 [Gossypium raimondii] |
| 15 |
Hb_000914_110 |
0.1742860253 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein WIP2 [Jatropha curcas] |
| 16 |
Hb_033053_020 |
0.1746752629 |
- |
- |
hypothetical protein JCGZ_01204 [Jatropha curcas] |
| 17 |
Hb_000009_300 |
0.1746761608 |
- |
- |
hypothetical protein CICLE_v10024731mg [Citrus clementina] |
| 18 |
Hb_000062_490 |
0.1783666365 |
- |
- |
PREDICTED: lipoxygenase 6, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001675_310 |
0.1789585711 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_020967_010 |
0.1804301431 |
- |
- |
- |