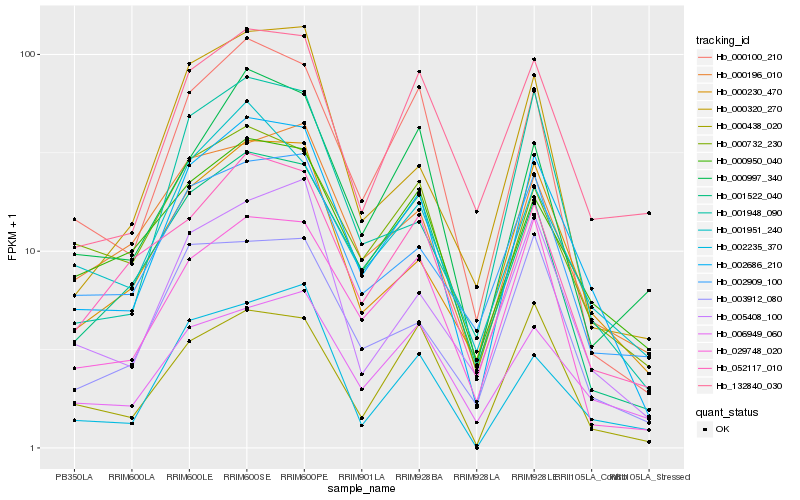
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_210 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_000230_470 |
0.1082818517 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000100_210 |
0.1143581056 |
- |
- |
PREDICTED: selenium-binding protein 1-like [Jatropha curcas] |
| 4 |
Hb_005408_100 |
0.1199391369 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 14 [Jatropha curcas] |
| 5 |
Hb_001522_040 |
0.1250873401 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_029748_020 |
0.1272307219 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000196_010 |
0.1283284513 |
- |
- |
Patellin-5, putative [Ricinus communis] |
| 8 |
Hb_003912_080 |
0.1287156391 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 9 |
Hb_006949_060 |
0.1291963165 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 10 |
Hb_052117_010 |
0.1305890647 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_002909_100 |
0.1334924678 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 12 |
Hb_001948_090 |
0.1340465649 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 13 |
Hb_000950_040 |
0.1340923495 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 14 |
Hb_000732_230 |
0.1341680071 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001951_240 |
0.1353867229 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 16 |
Hb_132840_030 |
0.1361308155 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |
| 17 |
Hb_000438_020 |
0.1363319067 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 18 |
Hb_002235_370 |
0.1368860657 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 19 |
Hb_000320_270 |
0.1389889658 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000997_340 |
0.1403298375 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |