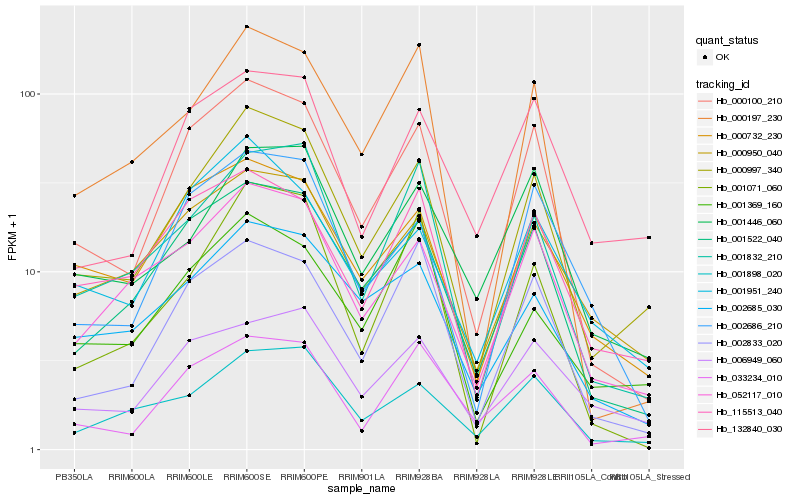
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000997_340 |
0.0 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 2 |
Hb_000100_210 |
0.1043946089 |
- |
- |
PREDICTED: selenium-binding protein 1-like [Jatropha curcas] |
| 3 |
Hb_052117_010 |
0.1062837605 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_001522_040 |
0.1129722435 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_001898_020 |
0.1156773491 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 6 |
Hb_001832_210 |
0.1172505801 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 7 |
Hb_000950_040 |
0.1223412828 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 8 |
Hb_002685_030 |
0.1272454962 |
- |
- |
PREDICTED: protein SMG7L [Jatropha curcas] |
| 9 |
Hb_000732_230 |
0.1287760384 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001446_060 |
0.1326557495 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_115513_040 |
0.1336052572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_132840_030 |
0.1352596318 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |
| 13 |
Hb_000197_230 |
0.1357026794 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 14 |
Hb_033234_010 |
0.1363570237 |
desease resistance |
Gene Name: LRR_8 |
Disease resistance protein RPP8 [Theobroma cacao] |
| 15 |
Hb_001951_240 |
0.1367786061 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 16 |
Hb_001071_060 |
0.1375795585 |
- |
- |
Trehalose phosphatase/synthase 5 isoform 1 [Theobroma cacao] |
| 17 |
Hb_002686_210 |
0.1403298375 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 18 |
Hb_002833_020 |
0.1404485808 |
desease resistance |
Gene Name: LRR_4 |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 19 |
Hb_006949_060 |
0.1407364766 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 20 |
Hb_001369_160 |
0.1418613926 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |