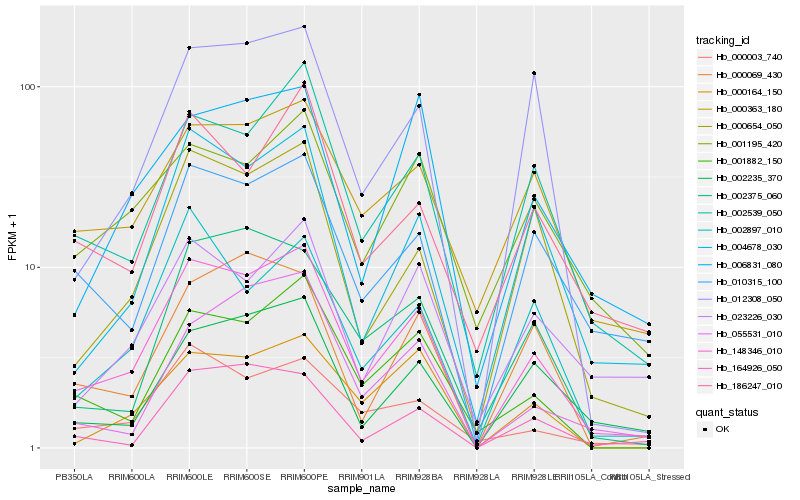
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148346_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_002539_050 |
0.1273064265 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 3 |
Hb_004678_030 |
0.1306131505 |
- |
- |
fructokinase [Manihot esculenta] |
| 4 |
Hb_000654_050 |
0.1358597486 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 5 |
Hb_010315_100 |
0.1389664181 |
- |
- |
PREDICTED: uncharacterized protein LOC104420162 [Eucalyptus grandis] |
| 6 |
Hb_000003_740 |
0.1409688415 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 7 |
Hb_001882_150 |
0.1417256145 |
transcription factor |
TF Family: mTERF |
- |
| 8 |
Hb_002235_370 |
0.1432029057 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 9 |
Hb_000363_180 |
0.1484637649 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 10 |
Hb_012308_050 |
0.1556456322 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 11 |
Hb_002897_010 |
0.1577769339 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g58300 [Jatropha curcas] |
| 12 |
Hb_186247_010 |
0.158614334 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase-like [Pyrus x bretschneideri] |
| 13 |
Hb_000164_150 |
0.1588149103 |
- |
- |
serine/threonine-specific protein kinase, putative [Ricinus communis] |
| 14 |
Hb_164926_050 |
0.1594670685 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 15 |
Hb_023226_030 |
0.15958589 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-14-like [Jatropha curcas] |
| 16 |
Hb_002375_060 |
0.1605240484 |
- |
- |
PREDICTED: uncharacterized protein LOC105628271 [Jatropha curcas] |
| 17 |
Hb_000069_430 |
0.1609850442 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 18 |
Hb_006831_080 |
0.1653122735 |
- |
- |
PREDICTED: uncharacterized protein LOC105642126 [Jatropha curcas] |
| 19 |
Hb_001195_420 |
0.1660952992 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X4 [Jatropha curcas] |
| 20 |
Hb_055531_010 |
0.1664769684 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Pyrus x bretschneideri] |