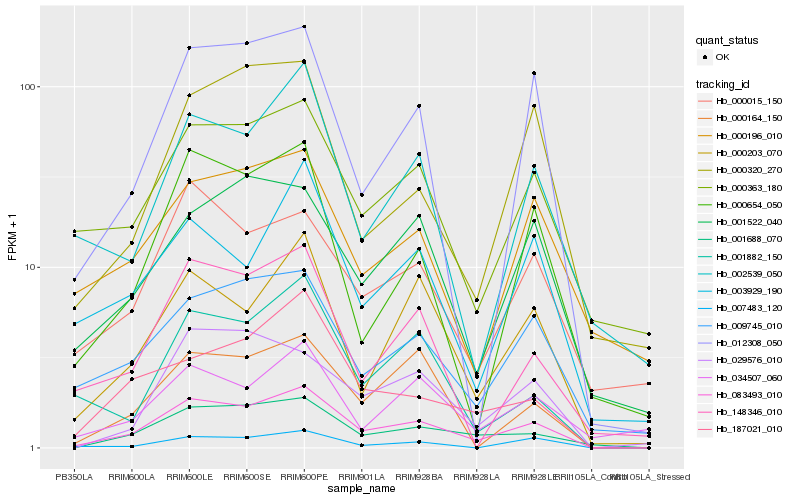
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_083493_010 |
0.0 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 2 |
Hb_001688_070 |
0.1383819029 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X1 [Jatropha curcas] |
| 3 |
Hb_009745_010 |
0.1498448627 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 4 |
Hb_000203_070 |
0.1523826864 |
- |
- |
PREDICTED: uncharacterized protein LOC105649674 [Jatropha curcas] |
| 5 |
Hb_012308_050 |
0.1536737736 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 6 |
Hb_000363_180 |
0.1600320671 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 7 |
Hb_187021_010 |
0.1647932791 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 8 |
Hb_007483_120 |
0.1668494823 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 9 |
Hb_000654_050 |
0.168733027 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 10 |
Hb_000015_150 |
0.1690639396 |
- |
- |
Ubiquitin-conjugating enzyme E2 7 family protein [Populus trichocarpa] |
| 11 |
Hb_000164_150 |
0.1692504465 |
- |
- |
serine/threonine-specific protein kinase, putative [Ricinus communis] |
| 12 |
Hb_148346_010 |
0.1697788551 |
- |
- |
- |
| 13 |
Hb_029576_010 |
0.1708766865 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31-like [Glycine max] |
| 14 |
Hb_001522_040 |
0.171397125 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_001882_150 |
0.1721771223 |
transcription factor |
TF Family: mTERF |
- |
| 16 |
Hb_002539_050 |
0.1752017302 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 17 |
Hb_003929_190 |
0.1795693934 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g18500 [Jatropha curcas] |
| 18 |
Hb_000320_270 |
0.1836430692 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000196_010 |
0.1837131671 |
- |
- |
Patellin-5, putative [Ricinus communis] |
| 20 |
Hb_034507_060 |
0.1848109075 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic-like [Citrus sinensis] |