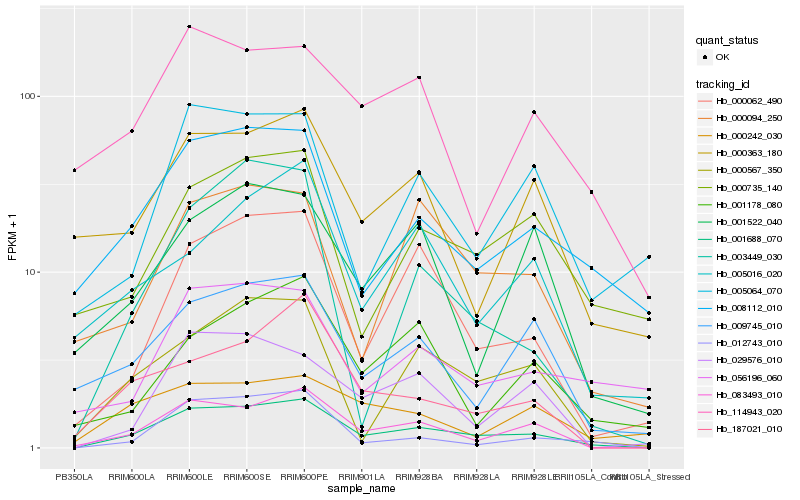
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001688_070 |
0.0 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X1 [Jatropha curcas] |
| 2 |
Hb_083493_010 |
0.1383819029 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 3 |
Hb_000062_490 |
0.1745155077 |
- |
- |
PREDICTED: lipoxygenase 6, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000567_350 |
0.1807798903 |
- |
- |
PREDICTED: probable acyl-activating enzyme 6 [Jatropha curcas] |
| 5 |
Hb_008112_010 |
0.1818016088 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 6 |
Hb_187021_010 |
0.1853730911 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 7 |
Hb_012743_010 |
0.188847235 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 8 |
Hb_029576_010 |
0.1889704497 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31-like [Glycine max] |
| 9 |
Hb_009745_010 |
0.1900621284 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 10 |
Hb_000094_250 |
0.1998806711 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X3 [Jatropha curcas] |
| 11 |
Hb_000363_180 |
0.2036042105 |
- |
- |
PREDICTED: uncharacterized protein LOC105633052 [Jatropha curcas] |
| 12 |
Hb_005016_020 |
0.2047100065 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_000735_140 |
0.2061586533 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 14 |
Hb_001178_080 |
0.2063640107 |
- |
- |
PREDICTED: protein kinase PVPK-1 [Jatropha curcas] |
| 15 |
Hb_056196_060 |
0.2065039297 |
- |
- |
hexokinase [Manihot esculenta] |
| 16 |
Hb_001522_040 |
0.208409389 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_005064_070 |
0.2084703258 |
- |
- |
PREDICTED: uncharacterized protein LOC105648318 [Jatropha curcas] |
| 18 |
Hb_114943_020 |
0.208852914 |
- |
- |
PREDICTED: NADPH--cytochrome P450 reductase 2 [Jatropha curcas] |
| 19 |
Hb_003449_030 |
0.2101996653 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 3, chloroplastic-like [Camelina sativa] |
| 20 |
Hb_000242_030 |
0.2114089459 |
- |
- |
PREDICTED: uncharacterized protein LOC102608003 [Citrus sinensis] |