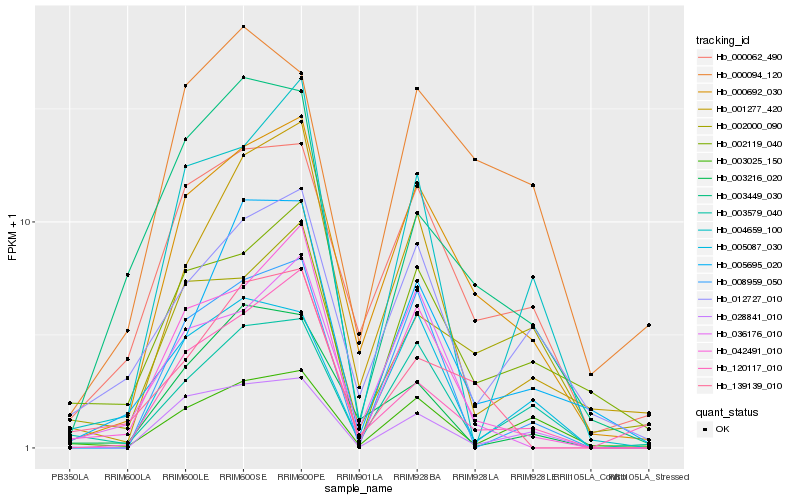
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000692_030 |
0.0 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 2 |
Hb_000062_490 |
0.1111356722 |
- |
- |
PREDICTED: lipoxygenase 6, chloroplastic [Jatropha curcas] |
| 3 |
Hb_042491_010 |
0.1554262982 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 4 |
Hb_001277_420 |
0.1667936635 |
- |
- |
PREDICTED: protein lin-12-like isoform X2 [Populus euphratica] |
| 5 |
Hb_012727_010 |
0.1675332509 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 6 |
Hb_028841_010 |
0.1727063039 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase CES101 [Jatropha curcas] |
| 7 |
Hb_003579_040 |
0.1772039346 |
- |
- |
PREDICTED: monoacylglycerol lipase ABHD6 [Jatropha curcas] |
| 8 |
Hb_120117_010 |
0.1791707302 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 9 |
Hb_036176_010 |
0.1816668319 |
- |
- |
hypothetical protein CISIN_1g002055mg [Citrus sinensis] |
| 10 |
Hb_002119_040 |
0.181828809 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 11 |
Hb_000094_120 |
0.1828539994 |
- |
- |
PREDICTED: uncharacterized protein At1g01500 [Jatropha curcas] |
| 12 |
Hb_003025_150 |
0.1861371889 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005695_020 |
0.1866096739 |
- |
- |
PREDICTED: low affinity sulfate transporter 3 [Jatropha curcas] |
| 14 |
Hb_139139_010 |
0.1895155969 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002000_090 |
0.1897445736 |
- |
- |
60S ribosomal protein L3B [Hevea brasiliensis] |
| 16 |
Hb_004659_100 |
0.1924299293 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005087_030 |
0.1932788194 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 18 |
Hb_003216_020 |
0.1936917712 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 19 |
Hb_003449_030 |
0.1939727693 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 3, chloroplastic-like [Camelina sativa] |
| 20 |
Hb_008959_050 |
0.1942585973 |
transcription factor |
TF Family: Orphans |
PREDICTED: late secretory pathway protein AVL9-like isoform X2 [Jatropha curcas] |