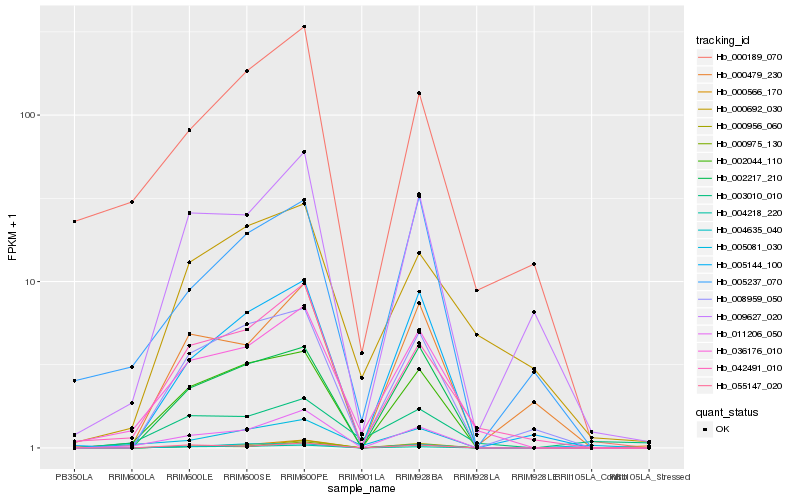
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_036176_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g002055mg [Citrus sinensis] |
| 2 |
Hb_042491_010 |
0.1290113081 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 3 |
Hb_011206_050 |
0.1466029458 |
- |
- |
hypothetical protein POPTR_0011s02840g [Populus trichocarpa] |
| 4 |
Hb_002044_110 |
0.1569389534 |
- |
- |
PREDICTED: probable carboxylesterase 5 isoform X1 [Populus euphratica] |
| 5 |
Hb_000189_070 |
0.1743106553 |
transcription factor |
TF Family: AUX/IAA |
Auxin-induced protein 22D, putative [Ricinus communis] |
| 6 |
Hb_002217_210 |
0.1783901205 |
- |
- |
- |
| 7 |
Hb_005144_100 |
0.1806643552 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 8 |
Hb_000692_030 |
0.1816668319 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 9 |
Hb_005237_070 |
0.1837453422 |
transcription factor |
TF Family: HB |
PREDICTED: WUSCHEL-related homeobox 4-like [Populus euphratica] |
| 10 |
Hb_009627_020 |
0.1854031998 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 11 |
Hb_004218_220 |
0.1878664071 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_005081_030 |
0.1899236346 |
- |
- |
hypothetical protein POPTR_0018s09200g [Populus trichocarpa] |
| 13 |
Hb_008959_050 |
0.1926424689 |
transcription factor |
TF Family: Orphans |
PREDICTED: late secretory pathway protein AVL9-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_055147_020 |
0.194892106 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 15 |
Hb_003010_010 |
0.1951360115 |
- |
- |
PREDICTED: uncharacterized protein LOC105633673 [Jatropha curcas] |
| 16 |
Hb_000566_170 |
0.1986059712 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL9 homolog isoform X4 [Jatropha curcas] |
| 17 |
Hb_000975_130 |
0.2035440538 |
- |
- |
PREDICTED: uncharacterized protein LOC104445138 [Eucalyptus grandis] |
| 18 |
Hb_004635_040 |
0.2051067239 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 19 |
Hb_000956_060 |
0.2052809407 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000479_230 |
0.2062417437 |
- |
- |
hypothetical protein JCGZ_19903 [Jatropha curcas] |