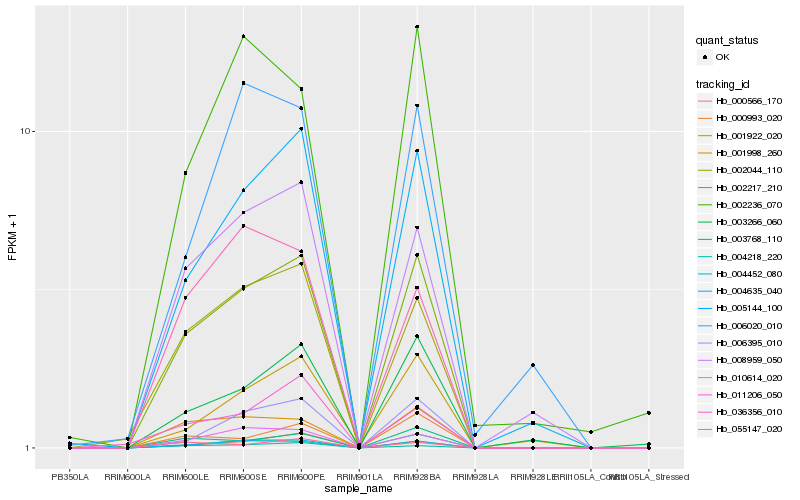
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_210 |
0.0 |
- |
- |
- |
| 2 |
Hb_004635_040 |
0.0901174416 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 3 |
Hb_005144_100 |
0.0941403147 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 4 |
Hb_002044_110 |
0.1032285141 |
- |
- |
PREDICTED: probable carboxylesterase 5 isoform X1 [Populus euphratica] |
| 5 |
Hb_001922_020 |
0.1063817619 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010614_020 |
0.1179394179 |
- |
- |
BnaC05g27570D [Brassica napus] |
| 7 |
Hb_008959_050 |
0.1279079872 |
transcription factor |
TF Family: Orphans |
PREDICTED: late secretory pathway protein AVL9-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_003768_110 |
0.1283482913 |
- |
- |
hypothetical protein POPTR_0004s12230g [Populus trichocarpa] |
| 9 |
Hb_004452_080 |
0.1361492694 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 10 |
Hb_002236_070 |
0.147974153 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |
| 11 |
Hb_006395_010 |
0.1480008458 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 12 |
Hb_003266_060 |
0.1493930204 |
- |
- |
PREDICTED: basic 7S globulin-like [Populus euphratica] |
| 13 |
Hb_004218_220 |
0.1520921373 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000993_020 |
0.1538043795 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 15 |
Hb_036356_010 |
0.1581263225 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_011206_050 |
0.1614178512 |
- |
- |
hypothetical protein POPTR_0011s02840g [Populus trichocarpa] |
| 17 |
Hb_000566_170 |
0.1636644824 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL9 homolog isoform X4 [Jatropha curcas] |
| 18 |
Hb_055147_020 |
0.168036483 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 19 |
Hb_006020_010 |
0.1687272527 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 20 |
Hb_001998_260 |
0.1689879643 |
- |
- |
cytochrome P450, putative [Ricinus communis] |