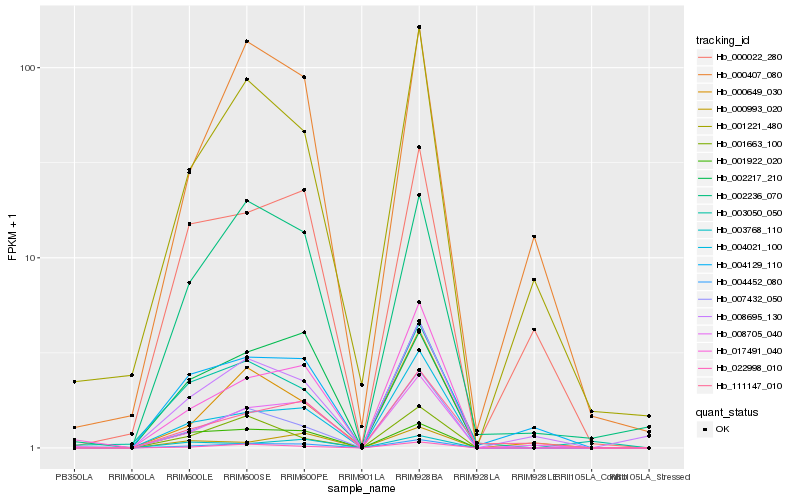
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004452_080 |
0.0 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 2 |
Hb_008705_040 |
0.1108214987 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 3 |
Hb_022998_010 |
0.1226256932 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_004021_100 |
0.1283000706 |
- |
- |
- |
| 5 |
Hb_007432_050 |
0.1306400964 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108 [Jatropha curcas] |
| 6 |
Hb_017491_040 |
0.1315807951 |
- |
- |
PREDICTED: uncharacterized protein LOC105631348 [Jatropha curcas] |
| 7 |
Hb_002236_070 |
0.1356289637 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |
| 8 |
Hb_002217_210 |
0.1361492694 |
- |
- |
- |
| 9 |
Hb_001922_020 |
0.139921779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008695_130 |
0.150412888 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003768_110 |
0.1526278177 |
- |
- |
hypothetical protein POPTR_0004s12230g [Populus trichocarpa] |
| 12 |
Hb_004129_110 |
0.1535484832 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 13 |
Hb_111147_010 |
0.1539109265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000649_030 |
0.155114128 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 15 |
Hb_001221_480 |
0.1566877892 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 16 |
Hb_000993_020 |
0.1598285658 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 17 |
Hb_003050_050 |
0.1634745834 |
- |
- |
hypothetical protein JCGZ_04619 [Jatropha curcas] |
| 18 |
Hb_000407_080 |
0.1636557426 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase [Hevea brasiliensis] |
| 19 |
Hb_001663_100 |
0.1645929821 |
- |
- |
- |
| 20 |
Hb_000022_280 |
0.1656963008 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |