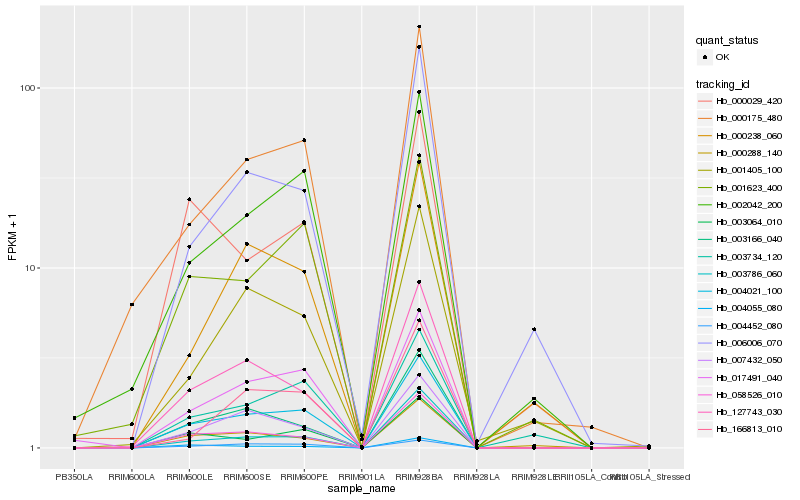
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004021_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_017491_040 |
0.0956460461 |
- |
- |
PREDICTED: uncharacterized protein LOC105631348 [Jatropha curcas] |
| 3 |
Hb_058526_010 |
0.0986470625 |
- |
- |
- |
| 4 |
Hb_003064_010 |
0.1047035506 |
- |
- |
Senescence-related gene 1 [Theobroma cacao] |
| 5 |
Hb_127743_030 |
0.1049676323 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_007432_050 |
0.1099024897 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108 [Jatropha curcas] |
| 7 |
Hb_003166_040 |
0.1117839291 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 8 |
Hb_001623_400 |
0.1146224498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002042_200 |
0.116781972 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_000175_480 |
0.1280003907 |
- |
- |
hypothetical protein JCGZ_06165 [Jatropha curcas] |
| 11 |
Hb_004452_080 |
0.1283000706 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 12 |
Hb_003786_060 |
0.1322441905 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 13 |
Hb_000288_140 |
0.1400919566 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 35 [Jatropha curcas] |
| 14 |
Hb_003734_120 |
0.1416898747 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006006_070 |
0.1417524363 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000238_060 |
0.1437721226 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 17 |
Hb_001405_100 |
0.144098328 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 18 |
Hb_004055_080 |
0.1451851311 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 19 |
Hb_166813_010 |
0.1464665441 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 20 |
Hb_000029_420 |
0.1496487338 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |