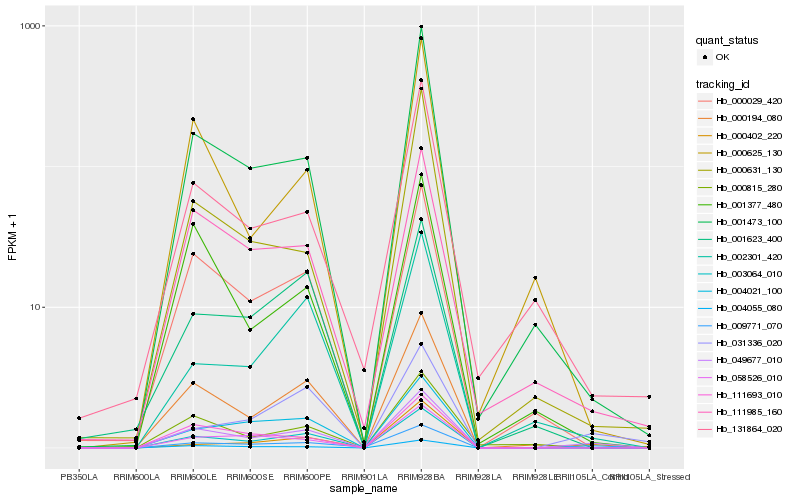
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_049677_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 2 |
Hb_009771_070 |
0.1150445818 |
- |
- |
hypothetical protein POPTR_0016s12140g [Populus trichocarpa] |
| 3 |
Hb_003064_010 |
0.1341085457 |
- |
- |
Senescence-related gene 1 [Theobroma cacao] |
| 4 |
Hb_000194_080 |
0.1453746839 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 5 |
Hb_004055_080 |
0.1473041823 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 6 |
Hb_000029_420 |
0.1483237435 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 7 |
Hb_001473_100 |
0.1533888048 |
- |
- |
chalcone synthase 1 [Jatropha curcas] |
| 8 |
Hb_111985_160 |
0.1563483791 |
- |
- |
hypothetical protein B456_010G008900 [Gossypium raimondii] |
| 9 |
Hb_111693_010 |
0.1658382791 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 10 |
Hb_058526_010 |
0.1720740371 |
- |
- |
- |
| 11 |
Hb_004021_100 |
0.1757449083 |
- |
- |
- |
| 12 |
Hb_000402_220 |
0.177792789 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 13 |
Hb_131864_020 |
0.1833985221 |
- |
- |
PREDICTED: anthocyanidin reductase [Jatropha curcas] |
| 14 |
Hb_001377_480 |
0.1856643305 |
transcription factor |
TF Family: MYB |
Myb domain protein 7, putative [Theobroma cacao] |
| 15 |
Hb_031336_020 |
0.1873032477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001623_400 |
0.1905365943 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002301_420 |
0.1909243697 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0011s05320g [Populus trichocarpa] |
| 18 |
Hb_000631_130 |
0.1911864685 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 19 |
Hb_000815_280 |
0.1912107168 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 3 [Jatropha curcas] |
| 20 |
Hb_000625_130 |
0.1914954946 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |