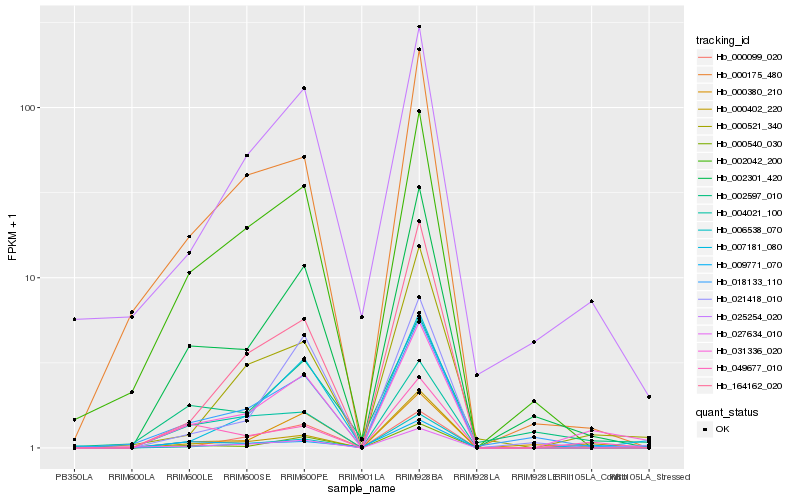
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031336_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002301_420 |
0.1622703417 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0011s05320g [Populus trichocarpa] |
| 3 |
Hb_000402_220 |
0.1652219581 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 4 |
Hb_000380_210 |
0.1678684771 |
- |
- |
Cyclic nucleotide-gated channel 15 [Theobroma cacao] |
| 5 |
Hb_000521_340 |
0.1741921287 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 6 |
Hb_025254_020 |
0.175688825 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Vitis vinifera] |
| 7 |
Hb_018133_110 |
0.1778438928 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_007181_080 |
0.1804663996 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000540_030 |
0.1833010478 |
- |
- |
Chitinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_049677_010 |
0.1873032477 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 11 |
Hb_002042_200 |
0.1906877777 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_027634_010 |
0.1973442489 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_006538_070 |
0.1975640345 |
- |
- |
hypothetical protein JCGZ_18339 [Jatropha curcas] |
| 14 |
Hb_002597_010 |
0.1978632049 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 15 |
Hb_009771_070 |
0.1984861074 |
- |
- |
hypothetical protein POPTR_0016s12140g [Populus trichocarpa] |
| 16 |
Hb_021418_010 |
0.1991063179 |
- |
- |
PREDICTED: probable pectate lyase 12 [Prunus mume] |
| 17 |
Hb_164162_020 |
0.1998793542 |
- |
- |
PREDICTED: uncharacterized protein LOC105646964 [Jatropha curcas] |
| 18 |
Hb_000099_020 |
0.2002174322 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: uncharacterized protein LOC103431057 [Malus domestica] |
| 19 |
Hb_004021_100 |
0.200535229 |
- |
- |
- |
| 20 |
Hb_000175_480 |
0.2009145179 |
- |
- |
hypothetical protein JCGZ_06165 [Jatropha curcas] |