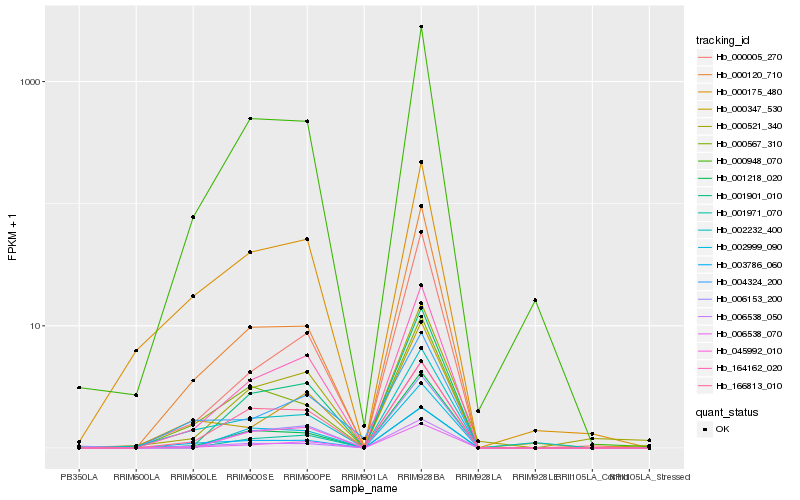
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006538_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_18339 [Jatropha curcas] |
| 2 |
Hb_164162_020 |
0.0302758317 |
- |
- |
PREDICTED: uncharacterized protein LOC105646964 [Jatropha curcas] |
| 3 |
Hb_001901_010 |
0.0897996481 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 4 |
Hb_000948_070 |
0.0990372426 |
- |
- |
hypothetical protein POPTR_0016s01720g [Populus trichocarpa] |
| 5 |
Hb_006538_050 |
0.0995315147 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 6 |
Hb_001971_070 |
0.1011883628 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_000005_270 |
0.1022130745 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 8 |
Hb_000120_710 |
0.1118787738 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_003786_060 |
0.1190744232 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 10 |
Hb_001218_020 |
0.126249265 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s17080g [Populus trichocarpa] |
| 11 |
Hb_000521_340 |
0.126868307 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 12 |
Hb_006153_200 |
0.1277700901 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein [Theobroma cacao] |
| 13 |
Hb_166813_010 |
0.1303303959 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 14 |
Hb_004324_200 |
0.1327634792 |
- |
- |
PREDICTED: transcription factor TRY-like [Populus euphratica] |
| 15 |
Hb_000347_530 |
0.1330222085 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB114-like [Jatropha curcas] |
| 16 |
Hb_045992_010 |
0.1334303429 |
- |
- |
- |
| 17 |
Hb_002232_400 |
0.1358643874 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH25-like [Jatropha curcas] |
| 18 |
Hb_000175_480 |
0.1387754513 |
- |
- |
hypothetical protein JCGZ_06165 [Jatropha curcas] |
| 19 |
Hb_002999_090 |
0.1406669984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000567_310 |
0.1410675709 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |