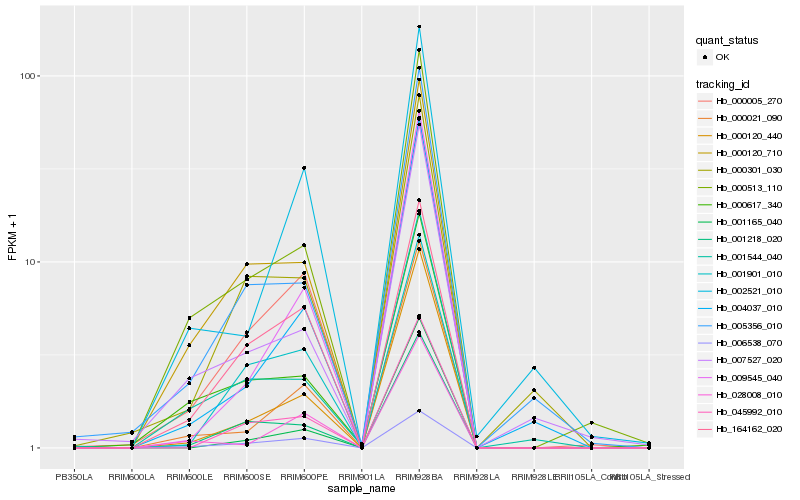
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000005_270 |
0.0 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 2 |
Hb_000120_440 |
0.0683952169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000513_110 |
0.0873491497 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Populus euphratica] |
| 4 |
Hb_045992_010 |
0.0878206842 |
- |
- |
- |
| 5 |
Hb_000021_090 |
0.0887579573 |
- |
- |
- |
| 6 |
Hb_000120_710 |
0.0897743074 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_009545_040 |
0.0973573177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006538_070 |
0.1022130745 |
- |
- |
hypothetical protein JCGZ_18339 [Jatropha curcas] |
| 9 |
Hb_001218_020 |
0.1080503285 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s17080g [Populus trichocarpa] |
| 10 |
Hb_000301_030 |
0.1083282613 |
- |
- |
PREDICTED: pectate lyase-like [Jatropha curcas] |
| 11 |
Hb_005356_010 |
0.1091742779 |
- |
- |
hypothetical protein POPTR_0003s16550g [Populus trichocarpa] |
| 12 |
Hb_004037_010 |
0.1128776179 |
- |
- |
PREDICTED: uncharacterized protein LOC105632369 [Jatropha curcas] |
| 13 |
Hb_001901_010 |
0.1141043078 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 14 |
Hb_000617_340 |
0.1153084738 |
- |
- |
respiratory burst oxidase B [Manihot esculenta] |
| 15 |
Hb_002521_010 |
0.1157352886 |
- |
- |
cinnamate 4-hydroxylase, putative [Ricinus communis] |
| 16 |
Hb_164162_020 |
0.1172041276 |
- |
- |
PREDICTED: uncharacterized protein LOC105646964 [Jatropha curcas] |
| 17 |
Hb_001544_040 |
0.1177314546 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 18 |
Hb_028008_010 |
0.1181532041 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 19 |
Hb_001165_040 |
0.1232615251 |
- |
- |
hypothetical protein POPTR_0009s10120g [Populus trichocarpa] |
| 20 |
Hb_007527_020 |
0.123543275 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |