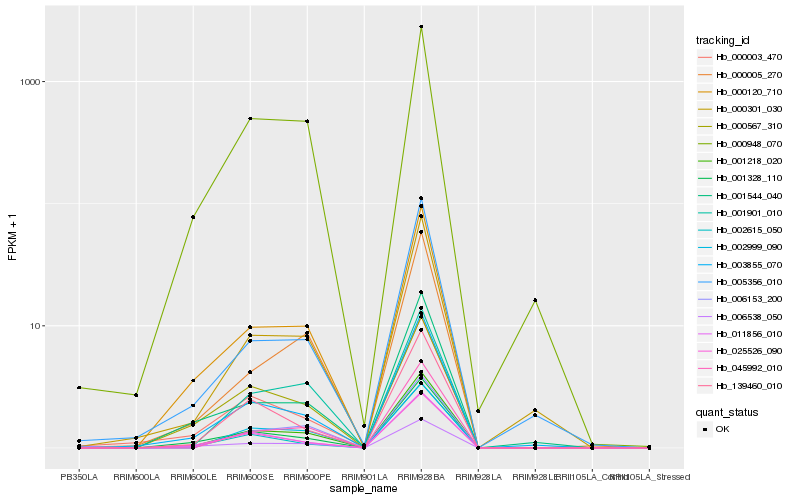
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001218_020 |
0.0 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s17080g [Populus trichocarpa] |
| 2 |
Hb_000120_710 |
0.0784221477 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_003855_070 |
0.0891643479 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 4 |
Hb_045992_010 |
0.0948713637 |
- |
- |
- |
| 5 |
Hb_001328_110 |
0.0977477409 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 6 |
Hb_006538_050 |
0.0977858393 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 7 |
Hb_000003_470 |
0.0982812797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006153_200 |
0.099336798 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein [Theobroma cacao] |
| 9 |
Hb_000301_030 |
0.0999524238 |
- |
- |
PREDICTED: pectate lyase-like [Jatropha curcas] |
| 10 |
Hb_001901_010 |
0.1007590311 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 11 |
Hb_011856_010 |
0.1024964573 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_000948_070 |
0.1060886511 |
- |
- |
hypothetical protein POPTR_0016s01720g [Populus trichocarpa] |
| 13 |
Hb_000005_270 |
0.1080503285 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 14 |
Hb_139460_010 |
0.1098105245 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 15 |
Hb_025526_090 |
0.1113477259 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 16 |
Hb_002999_090 |
0.1150250024 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001544_040 |
0.1188558613 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 18 |
Hb_002615_050 |
0.1191736969 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 19 |
Hb_000567_310 |
0.1194303746 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 20 |
Hb_005356_010 |
0.1196400337 |
- |
- |
hypothetical protein POPTR_0003s16550g [Populus trichocarpa] |