| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
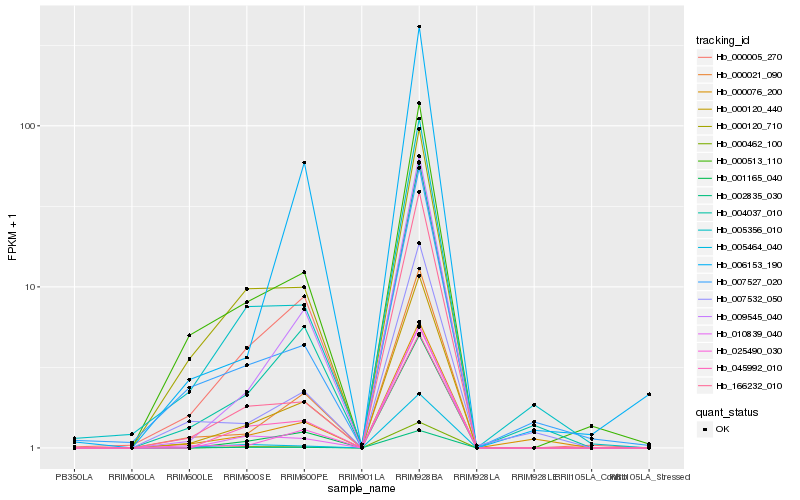
Hb_000120_440 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_009545_040 |
0.0520038408 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000021_090 |
0.0543234402 |
- |
- |
- |
| 4 |
Hb_001165_040 |
0.0661869532 |
- |
- |
hypothetical protein POPTR_0009s10120g [Populus trichocarpa] |
| 5 |
Hb_004037_010 |
0.0674954899 |
- |
- |
PREDICTED: uncharacterized protein LOC105632369 [Jatropha curcas] |
| 6 |
Hb_000005_270 |
0.0683952169 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 7 |
Hb_000513_110 |
0.0853499646 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Populus euphratica] |
| 8 |
Hb_025490_030 |
0.0870216553 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |
| 9 |
Hb_005356_010 |
0.0961833783 |
- |
- |
hypothetical protein POPTR_0003s16550g [Populus trichocarpa] |
| 10 |
Hb_045992_010 |
0.1004544149 |
- |
- |
- |
| 11 |
Hb_000462_100 |
0.1067068217 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 12 |
Hb_000120_710 |
0.10745281 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_002835_030 |
0.1094324609 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Vitis vinifera] |
| 14 |
Hb_010839_040 |
0.109678672 |
- |
- |
hypothetical protein JCGZ_20868 [Jatropha curcas] |
| 15 |
Hb_000076_200 |
0.1105399541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007527_020 |
0.1108904451 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |
| 17 |
Hb_006153_190 |
0.1135174068 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Gossypium raimondii] |
| 18 |
Hb_007532_050 |
0.1135624074 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 19 |
Hb_005464_040 |
0.1139629784 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_166232_010 |
0.1140325152 |
- |
- |
hypothetical protein JCGZ_22831 [Jatropha curcas] |