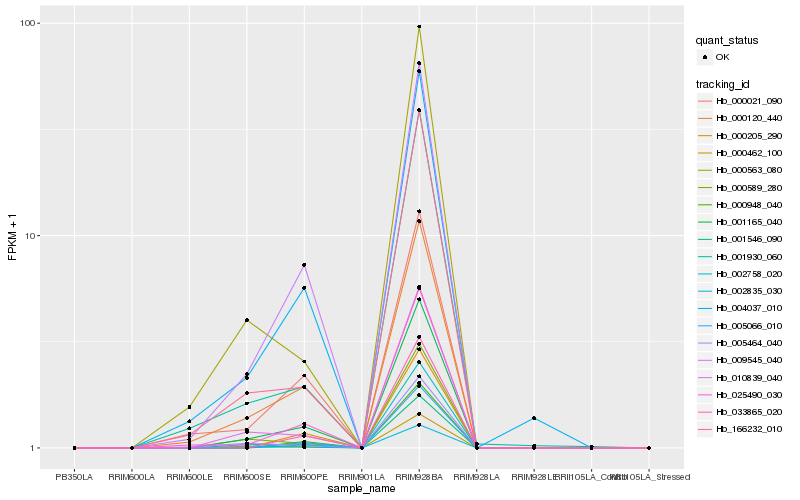
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001165_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s10120g [Populus trichocarpa] |
| 2 |
Hb_009545_040 |
0.0558167689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000120_440 |
0.0661869532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002835_030 |
0.0665514327 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Vitis vinifera] |
| 5 |
Hb_000462_100 |
0.0713573219 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 6 |
Hb_010839_040 |
0.0725677793 |
- |
- |
hypothetical protein JCGZ_20868 [Jatropha curcas] |
| 7 |
Hb_004037_010 |
0.0792348368 |
- |
- |
PREDICTED: uncharacterized protein LOC105632369 [Jatropha curcas] |
| 8 |
Hb_002758_020 |
0.0795351339 |
- |
- |
PREDICTED: putative auxin efflux carrier component 5 [Jatropha curcas] |
| 9 |
Hb_025490_030 |
0.081086752 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |
| 10 |
Hb_166232_010 |
0.0889031523 |
- |
- |
hypothetical protein JCGZ_22831 [Jatropha curcas] |
| 11 |
Hb_000021_090 |
0.0929762167 |
- |
- |
- |
| 12 |
Hb_000589_280 |
0.0949538158 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |
| 13 |
Hb_001930_060 |
0.1016785182 |
- |
- |
PREDICTED: cytochrome P450 71D9-like [Fragaria vesca subsp. vesca] |
| 14 |
Hb_005066_010 |
0.1073924136 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 15 |
Hb_033865_020 |
0.1075759696 |
- |
- |
- |
| 16 |
Hb_000948_040 |
0.1076643629 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Jatropha curcas] |
| 17 |
Hb_005464_040 |
0.1098457901 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000205_290 |
0.1132125792 |
- |
- |
- |
| 19 |
Hb_001546_090 |
0.113892408 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB114-like [Jatropha curcas] |
| 20 |
Hb_000563_080 |
0.1147020156 |
- |
- |
- |