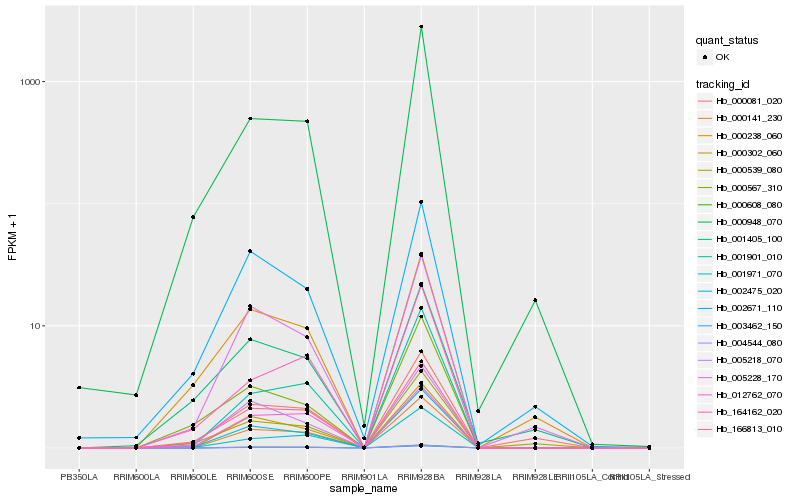
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_166813_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 2 |
Hb_002475_020 |
0.0882331322 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X1 [Populus euphratica] |
| 3 |
Hb_000948_070 |
0.0957466029 |
- |
- |
hypothetical protein POPTR_0016s01720g [Populus trichocarpa] |
| 4 |
Hb_000608_080 |
0.0984733804 |
- |
- |
hypothetical protein JCGZ_22216 [Jatropha curcas] |
| 5 |
Hb_000141_230 |
0.1013911503 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Camelina sativa] |
| 6 |
Hb_004544_080 |
0.1014520742 |
- |
- |
lectin protein kinase [Populus trichocarpa] |
| 7 |
Hb_000238_060 |
0.1016035504 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 8 |
Hb_003462_150 |
0.1020439742 |
- |
- |
PREDICTED: chitinase 1-like [Eucalyptus grandis] |
| 9 |
Hb_012762_070 |
0.1029322828 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HOX3 [Jatropha curcas] |
| 10 |
Hb_005218_070 |
0.1053707197 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX3 isoform X2 [Populus euphratica] |
| 11 |
Hb_005228_170 |
0.105406446 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 12 |
Hb_164162_020 |
0.1069791688 |
- |
- |
PREDICTED: uncharacterized protein LOC105646964 [Jatropha curcas] |
| 13 |
Hb_001971_070 |
0.1086049788 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_002671_110 |
0.1126771001 |
- |
- |
PREDICTED: remorin-like [Jatropha curcas] |
| 15 |
Hb_001405_100 |
0.1187670528 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 16 |
Hb_000081_020 |
0.1188468367 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 1.1-like [Camelina sativa] |
| 17 |
Hb_000567_310 |
0.1199192998 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |
| 18 |
Hb_000302_060 |
0.1204590182 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_000539_080 |
0.1224982041 |
- |
- |
hypothetical protein POPTR_0039s00260g, partial [Populus trichocarpa] |
| 20 |
Hb_001901_010 |
0.1252069497 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |