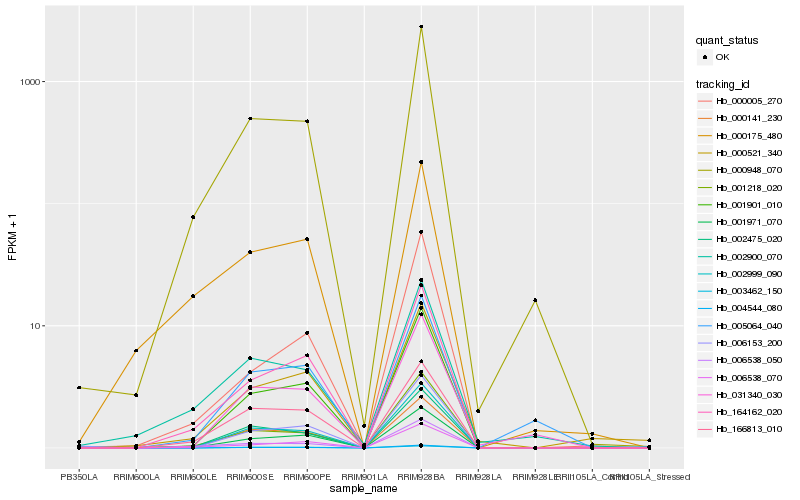
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000521_340 |
0.0 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 2 |
Hb_001971_070 |
0.1170281722 |
- |
- |
unnamed protein product [Coffea canephora] |
| 3 |
Hb_164162_020 |
0.1172571358 |
- |
- |
PREDICTED: uncharacterized protein LOC105646964 [Jatropha curcas] |
| 4 |
Hb_001901_010 |
0.1248387429 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 5 |
Hb_006538_070 |
0.126868307 |
- |
- |
hypothetical protein JCGZ_18339 [Jatropha curcas] |
| 6 |
Hb_000948_070 |
0.1339747314 |
- |
- |
hypothetical protein POPTR_0016s01720g [Populus trichocarpa] |
| 7 |
Hb_166813_010 |
0.1478219071 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 8 |
Hb_004544_080 |
0.1486871262 |
- |
- |
lectin protein kinase [Populus trichocarpa] |
| 9 |
Hb_002999_090 |
0.1490069226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003462_150 |
0.1492508755 |
- |
- |
PREDICTED: chitinase 1-like [Eucalyptus grandis] |
| 11 |
Hb_006153_200 |
0.1530714916 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein [Theobroma cacao] |
| 12 |
Hb_002475_020 |
0.1545524872 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X1 [Populus euphratica] |
| 13 |
Hb_002900_070 |
0.1569532454 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_031340_030 |
0.1571044966 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Jatropha curcas] |
| 15 |
Hb_000141_230 |
0.1584188136 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Camelina sativa] |
| 16 |
Hb_000005_270 |
0.1607865782 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 17 |
Hb_006538_050 |
0.1623095776 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 18 |
Hb_001218_020 |
0.1635012799 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s17080g [Populus trichocarpa] |
| 19 |
Hb_005064_040 |
0.1640488361 |
- |
- |
hypothetical protein POPTR_0008s02680g [Populus trichocarpa] |
| 20 |
Hb_000175_480 |
0.1644650173 |
- |
- |
hypothetical protein JCGZ_06165 [Jatropha curcas] |