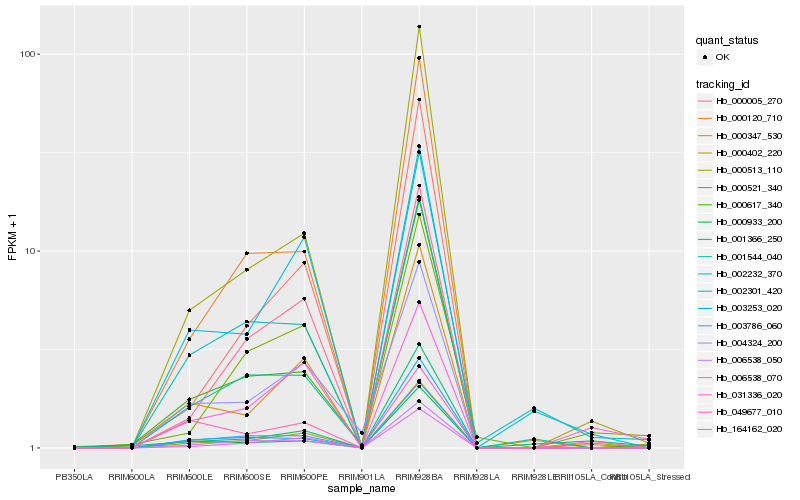
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000402_220 |
0.0 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 2 |
Hb_000513_110 |
0.1486480327 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Populus euphratica] |
| 3 |
Hb_006538_070 |
0.1531419097 |
- |
- |
hypothetical protein JCGZ_18339 [Jatropha curcas] |
| 4 |
Hb_006538_050 |
0.1541444441 |
- |
- |
hypothetical protein RCOM_1308310 [Ricinus communis] |
| 5 |
Hb_001366_250 |
0.1568012568 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 4-like [Jatropha curcas] |
| 6 |
Hb_000120_710 |
0.1590190769 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_003786_060 |
0.159309058 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 8 |
Hb_000005_270 |
0.1611703876 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 9 |
Hb_002232_370 |
0.1642309263 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 10 |
Hb_031336_020 |
0.1652219581 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000521_340 |
0.1658237054 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 12 |
Hb_000617_340 |
0.1665439492 |
- |
- |
respiratory burst oxidase B [Manihot esculenta] |
| 13 |
Hb_164162_020 |
0.1665865911 |
- |
- |
PREDICTED: uncharacterized protein LOC105646964 [Jatropha curcas] |
| 14 |
Hb_000933_200 |
0.1666121042 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 15 |
Hb_000347_530 |
0.1686909946 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB114-like [Jatropha curcas] |
| 16 |
Hb_001544_040 |
0.1745342295 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 17 |
Hb_002301_420 |
0.1755420743 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0011s05320g [Populus trichocarpa] |
| 18 |
Hb_003253_020 |
0.1759738968 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_049677_010 |
0.177792789 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 20 |
Hb_004324_200 |
0.1785220969 |
- |
- |
PREDICTED: transcription factor TRY-like [Populus euphratica] |