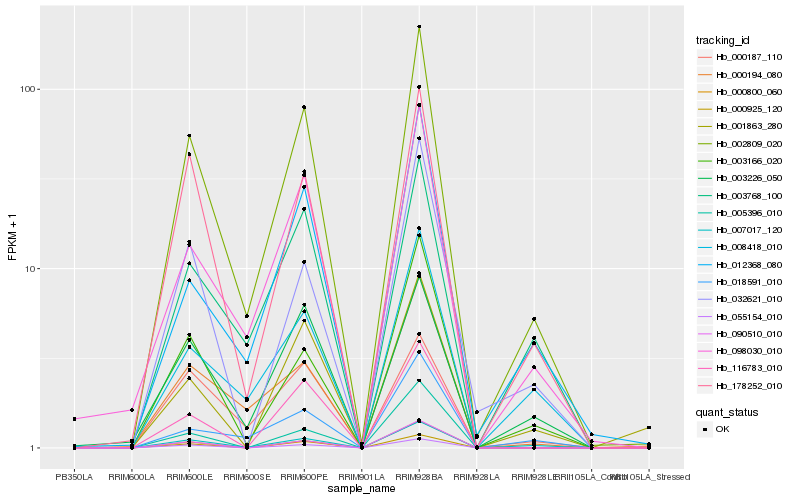
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002809_020 |
0.0 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 2 |
Hb_007017_120 |
0.0977792966 |
- |
- |
PREDICTED: exocyst complex component EXO84A [Jatropha curcas] |
| 3 |
Hb_178252_010 |
0.10206862 |
- |
- |
hypothetical protein POPTR_0018s01890g [Populus trichocarpa] |
| 4 |
Hb_098030_010 |
0.1050269802 |
- |
- |
hypothetical protein JCGZ_06251 [Jatropha curcas] |
| 5 |
Hb_055154_010 |
0.1183272499 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 6 |
Hb_003166_020 |
0.1207493047 |
- |
- |
PREDICTED: RNA-binding E3 ubiquitin-protein ligase MEX3C-like [Jatropha curcas] |
| 7 |
Hb_008418_010 |
0.1230902352 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |
| 8 |
Hb_000194_080 |
0.123154464 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 9 |
Hb_003768_100 |
0.1242568297 |
- |
- |
hypothetical protein JCGZ_19930 [Jatropha curcas] |
| 10 |
Hb_012368_080 |
0.1295519765 |
- |
- |
PREDICTED: uncharacterized protein LOC105126277 [Populus euphratica] |
| 11 |
Hb_090510_010 |
0.1314786107 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 12 |
Hb_000925_120 |
0.1315775881 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 13 |
Hb_116783_010 |
0.1345751816 |
- |
- |
hypothetical protein POPTR_0011s16190g [Populus trichocarpa] |
| 14 |
Hb_032621_010 |
0.1386305073 |
- |
- |
hypothetical protein PHAVU_006G197300g [Phaseolus vulgaris] |
| 15 |
Hb_000800_060 |
0.140005223 |
- |
- |
hypothetical protein POPTR_0008s02870g [Populus trichocarpa] |
| 16 |
Hb_018591_010 |
0.1401382478 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_003226_050 |
0.1410930944 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 18 |
Hb_005396_010 |
0.151882022 |
- |
- |
PREDICTED: interleukin enhancer-binding factor 3-B-like [Jatropha curcas] |
| 19 |
Hb_000187_110 |
0.1546385913 |
- |
- |
PREDICTED: serine carboxypeptidase-like 7 [Jatropha curcas] |
| 20 |
Hb_001863_280 |
0.1546620181 |
- |
- |
PREDICTED: protein EXORDIUM-like 2 [Jatropha curcas] |