| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
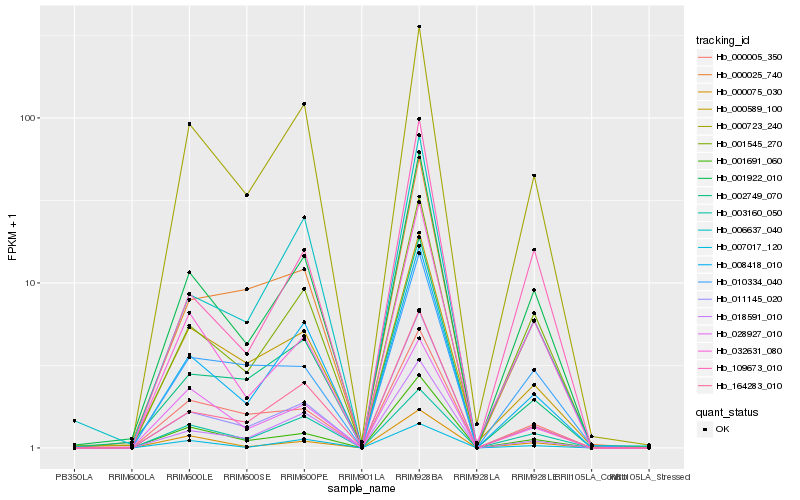
Hb_001922_010 |
0.0 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 2 |
Hb_001545_270 |
0.0623719331 |
- |
- |
laccase, putative [Ricinus communis] |
| 3 |
Hb_008418_010 |
0.0890773315 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |
| 4 |
Hb_000723_240 |
0.0936125191 |
- |
- |
phenylalanine ammonia-lyase 2 [Manihot esculenta] |
| 5 |
Hb_032631_080 |
0.0960635504 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_001691_060 |
0.1015041206 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 7 |
Hb_011145_020 |
0.1021534425 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Pyrus x bretschneideri] |
| 8 |
Hb_164283_010 |
0.1065974742 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 9 |
Hb_000589_100 |
0.1126414884 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_000075_030 |
0.113527171 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C10-like [Fragaria vesca subsp. vesca] |
| 11 |
Hb_109673_010 |
0.1154281636 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 12 |
Hb_002749_070 |
0.1199242576 |
- |
- |
PREDICTED: uncharacterized protein LOC105635941 [Jatropha curcas] |
| 13 |
Hb_000005_350 |
0.1205016132 |
- |
- |
PREDICTED: uncharacterized protein LOC105634263 [Jatropha curcas] |
| 14 |
Hb_000025_740 |
0.122636257 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 15 |
Hb_018591_010 |
0.1253404396 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_003160_050 |
0.1255920527 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 17 |
Hb_028927_010 |
0.1258946038 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 18 |
Hb_006637_040 |
0.1261641041 |
- |
- |
phenylalanine ammonia-lyase 2 [Manihot esculenta] |
| 19 |
Hb_007017_120 |
0.1274538461 |
- |
- |
PREDICTED: exocyst complex component EXO84A [Jatropha curcas] |
| 20 |
Hb_010334_040 |
0.1280056839 |
- |
- |
PREDICTED: uncharacterized protein LOC105108579 [Populus euphratica] |