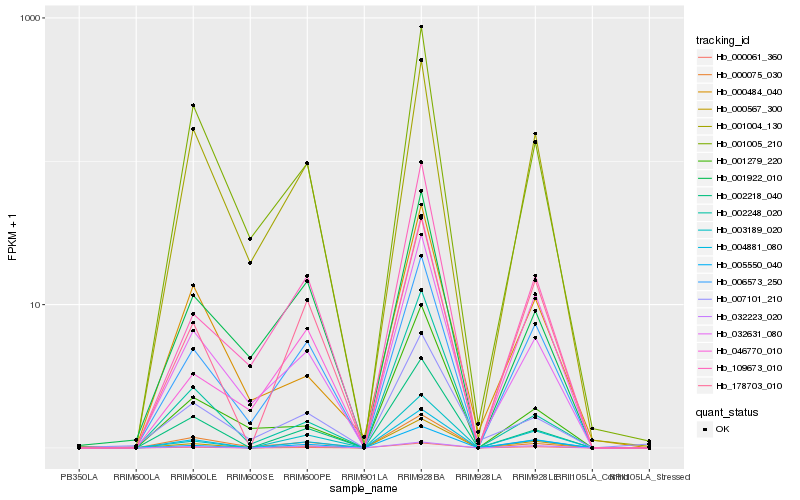
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004881_080 |
0.0 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 1 [Populus euphratica] |
| 2 |
Hb_002218_040 |
0.0641864261 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000061_360 |
0.0735582916 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 4 |
Hb_000567_300 |
0.0960762738 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 5 |
Hb_003189_020 |
0.1061792511 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 6 |
Hb_032631_080 |
0.1123481854 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_005550_040 |
0.127072117 |
- |
- |
PREDICTED: epoxide hydrolase 4-like [Jatropha curcas] |
| 8 |
Hb_000075_030 |
0.1278002217 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C10-like [Fragaria vesca subsp. vesca] |
| 9 |
Hb_001005_210 |
0.1318026179 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 10 |
Hb_109673_010 |
0.1360084058 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 11 |
Hb_006573_250 |
0.1364358556 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 12 |
Hb_178703_010 |
0.1394150963 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 13 |
Hb_002248_020 |
0.1454230403 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like isoform X1 [Populus euphratica] |
| 14 |
Hb_001922_010 |
0.1606995935 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 15 |
Hb_032223_020 |
0.1610020451 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_001279_220 |
0.1620401395 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_001004_130 |
0.1622426736 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 18 |
Hb_000484_040 |
0.1624590754 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_007101_210 |
0.1624717167 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_046770_010 |
0.1641484751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |