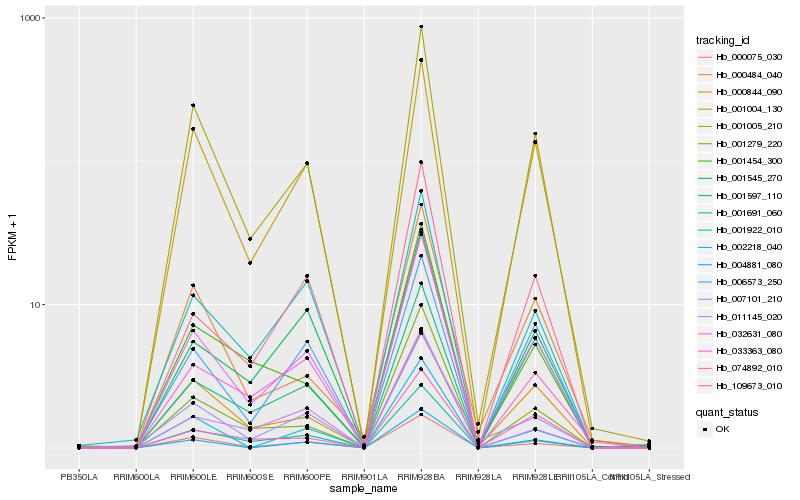
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032631_080 |
0.0 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_001005_210 |
0.0713460098 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 3 |
Hb_000075_030 |
0.0860787952 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C10-like [Fragaria vesca subsp. vesca] |
| 4 |
Hb_001922_010 |
0.0960635504 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 5 |
Hb_001691_060 |
0.1079714476 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 6 |
Hb_109673_010 |
0.1121069127 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 7 |
Hb_004881_080 |
0.1123481854 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 1 [Populus euphratica] |
| 8 |
Hb_006573_250 |
0.1139387388 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 9 |
Hb_007101_210 |
0.1147785767 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_001279_220 |
0.1162059319 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_001545_270 |
0.1172581348 |
- |
- |
laccase, putative [Ricinus communis] |
| 12 |
Hb_001004_130 |
0.1174085709 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 13 |
Hb_002218_040 |
0.1242319922 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000484_040 |
0.1253674431 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_001454_300 |
0.1271072321 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 16 |
Hb_033363_080 |
0.1289543602 |
- |
- |
Allene oxide cyclase 3, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_074892_010 |
0.1302650575 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 18 |
Hb_000844_090 |
0.1334058945 |
- |
- |
PREDICTED: cytochrome P450 82C4-like [Jatropha curcas] |
| 19 |
Hb_011145_020 |
0.1363826137 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Pyrus x bretschneideri] |
| 20 |
Hb_001597_110 |
0.137694609 |
- |
- |
- |