| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
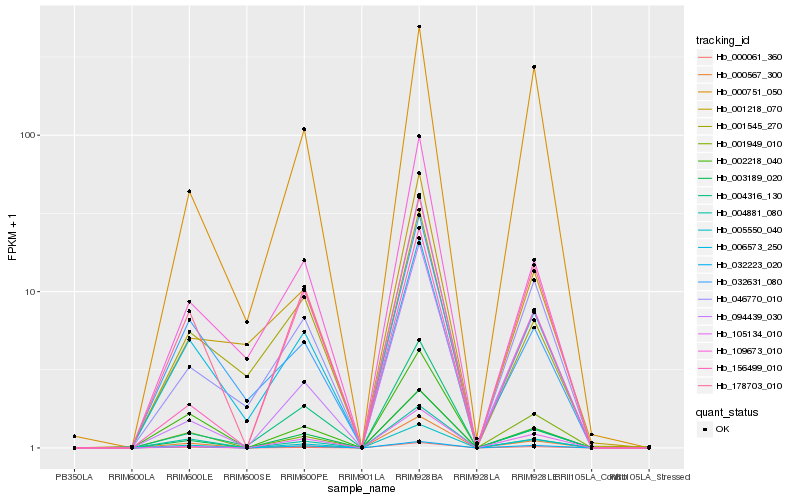
Hb_003189_020 |
0.0 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 2 |
Hb_005550_040 |
0.0608072579 |
- |
- |
PREDICTED: epoxide hydrolase 4-like [Jatropha curcas] |
| 3 |
Hb_105134_010 |
0.0911447824 |
- |
- |
hypothetical protein JCGZ_16347 [Jatropha curcas] |
| 4 |
Hb_178703_010 |
0.0943323015 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 5 |
Hb_046770_010 |
0.0974204983 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000061_360 |
0.1006800084 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 7 |
Hb_004881_080 |
0.1061792511 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 1 [Populus euphratica] |
| 8 |
Hb_109673_010 |
0.11659628 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 9 |
Hb_006573_250 |
0.1238190098 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 10 |
Hb_032223_020 |
0.1375576598 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_000567_300 |
0.1390833499 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 12 |
Hb_156499_010 |
0.1419270647 |
- |
- |
hypothetical protein JCGZ_21064 [Jatropha curcas] |
| 13 |
Hb_094439_030 |
0.1476576163 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g38780 [Eucalyptus grandis] |
| 14 |
Hb_001949_010 |
0.1480550162 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 15 |
Hb_001218_070 |
0.1487242142 |
- |
- |
PREDICTED: sugar transport protein 5-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_004316_130 |
0.1521257998 |
- |
- |
PREDICTED: glutathione transferase GST 23-like, partial [Eucalyptus grandis] |
| 17 |
Hb_000751_050 |
0.1570539773 |
- |
- |
PREDICTED: extensin, partial [Solanum lycopersicum] |
| 18 |
Hb_032631_080 |
0.1616608392 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_001545_270 |
0.1634384837 |
- |
- |
laccase, putative [Ricinus communis] |
| 20 |
Hb_002218_040 |
0.1634966029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |