| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
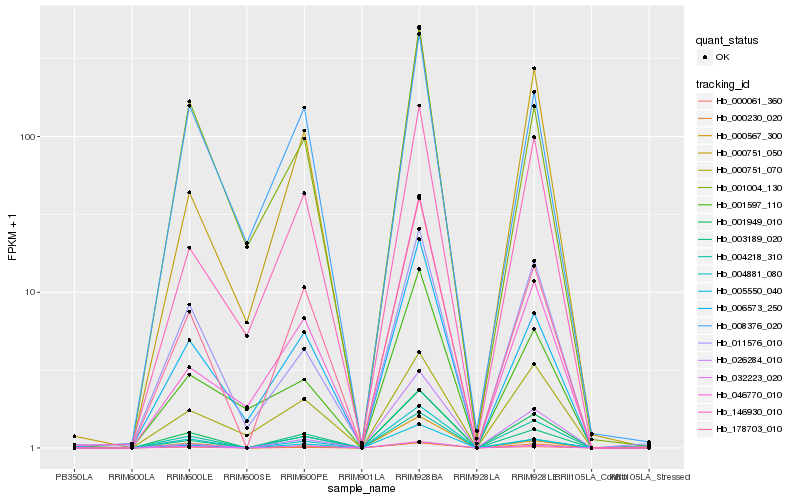
Hb_001949_010 |
0.0 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 2 |
Hb_005550_040 |
0.0956053468 |
- |
- |
PREDICTED: epoxide hydrolase 4-like [Jatropha curcas] |
| 3 |
Hb_011576_010 |
0.0999506105 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 4 |
Hb_178703_010 |
0.1039859908 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 5 |
Hb_032223_020 |
0.1224973293 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_000751_050 |
0.1243288625 |
- |
- |
PREDICTED: extensin, partial [Solanum lycopersicum] |
| 7 |
Hb_000061_360 |
0.1243350945 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 8 |
Hb_000230_020 |
0.1316399071 |
- |
- |
cysteine synthase, putative [Ricinus communis] |
| 9 |
Hb_006573_250 |
0.1345118595 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 10 |
Hb_001597_110 |
0.1411532733 |
- |
- |
- |
| 11 |
Hb_146930_010 |
0.1415411205 |
- |
- |
- |
| 12 |
Hb_003189_020 |
0.1480550162 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 13 |
Hb_004218_310 |
0.1631181181 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Camelina sativa] |
| 14 |
Hb_001004_130 |
0.1639242539 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 15 |
Hb_004881_080 |
0.1692211361 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 1 [Populus euphratica] |
| 16 |
Hb_000567_300 |
0.170889574 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 17 |
Hb_046770_010 |
0.1718315814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000751_070 |
0.1738479238 |
- |
- |
- |
| 19 |
Hb_026284_010 |
0.1759981347 |
- |
- |
- |
| 20 |
Hb_008376_020 |
0.1768296522 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |