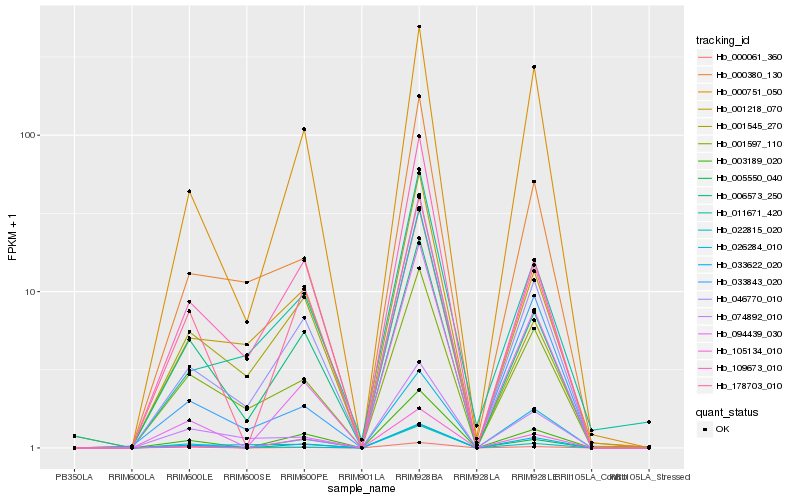
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_046770_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001218_070 |
0.0888260656 |
- |
- |
PREDICTED: sugar transport protein 5-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_109673_010 |
0.0920473194 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 4 |
Hb_000380_130 |
0.09410632 |
- |
- |
Phenylalanine ammonia-lyase, putative [Ricinus communis] |
| 5 |
Hb_003189_020 |
0.0974204983 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 6 |
Hb_005550_040 |
0.1006889169 |
- |
- |
PREDICTED: epoxide hydrolase 4-like [Jatropha curcas] |
| 7 |
Hb_105134_010 |
0.1008178498 |
- |
- |
hypothetical protein JCGZ_16347 [Jatropha curcas] |
| 8 |
Hb_011671_420 |
0.1120293551 |
- |
- |
hypothetical protein POPTR_0015s12340g [Populus trichocarpa] |
| 9 |
Hb_001597_110 |
0.1205627816 |
- |
- |
- |
| 10 |
Hb_094439_030 |
0.1261486594 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g38780 [Eucalyptus grandis] |
| 11 |
Hb_006573_250 |
0.129394372 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 12 |
Hb_074892_010 |
0.1343192649 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |
| 13 |
Hb_000751_050 |
0.136102594 |
- |
- |
PREDICTED: extensin, partial [Solanum lycopersicum] |
| 14 |
Hb_000061_360 |
0.139453516 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 15 |
Hb_033622_020 |
0.1403067661 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 16 |
Hb_026284_010 |
0.1436515665 |
- |
- |
- |
| 17 |
Hb_022815_020 |
0.1466623992 |
- |
- |
PREDICTED: uncharacterized protein LOC105650107 [Jatropha curcas] |
| 18 |
Hb_178703_010 |
0.1492441309 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 19 |
Hb_001545_270 |
0.154151494 |
- |
- |
laccase, putative [Ricinus communis] |
| 20 |
Hb_033843_020 |
0.1582891782 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |