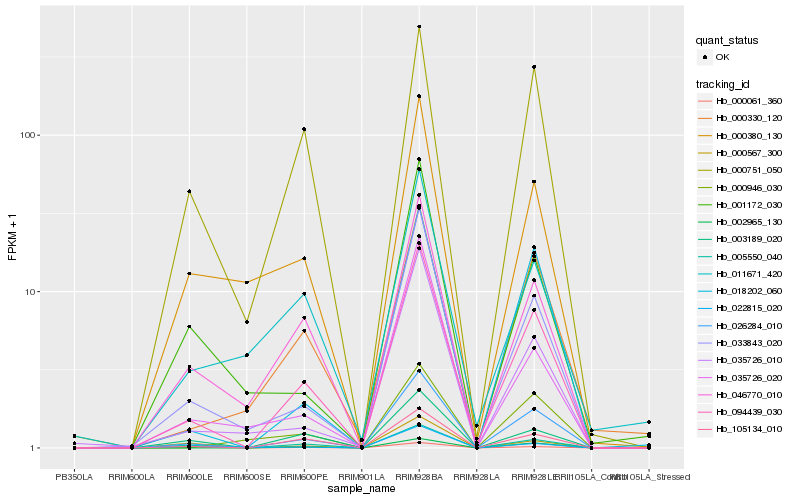
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_094439_030 |
0.0 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g38780 [Eucalyptus grandis] |
| 2 |
Hb_026284_010 |
0.0440610896 |
- |
- |
- |
| 3 |
Hb_105134_010 |
0.1049437582 |
- |
- |
hypothetical protein JCGZ_16347 [Jatropha curcas] |
| 4 |
Hb_033843_020 |
0.116593124 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 5 |
Hb_005550_040 |
0.1218968934 |
- |
- |
PREDICTED: epoxide hydrolase 4-like [Jatropha curcas] |
| 6 |
Hb_002965_130 |
0.1248634582 |
- |
- |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 7 |
Hb_046770_010 |
0.1261486594 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_018202_060 |
0.1410379722 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Jatropha curcas] |
| 9 |
Hb_000330_120 |
0.14176743 |
- |
- |
PREDICTED: bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase-like [Nelumbo nucifera] |
| 10 |
Hb_003189_020 |
0.1476576163 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 11 |
Hb_035726_010 |
0.1508680873 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 12 |
Hb_000751_050 |
0.1561854009 |
- |
- |
PREDICTED: extensin, partial [Solanum lycopersicum] |
| 13 |
Hb_000380_130 |
0.161175497 |
- |
- |
Phenylalanine ammonia-lyase, putative [Ricinus communis] |
| 14 |
Hb_000061_360 |
0.1628309287 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 15 |
Hb_035726_020 |
0.1680868083 |
- |
- |
Insulinase (Peptidase family M16) family protein [Theobroma cacao] |
| 16 |
Hb_000946_030 |
0.1717165216 |
- |
- |
hypothetical protein JCGZ_10707 [Jatropha curcas] |
| 17 |
Hb_011671_420 |
0.1736934889 |
- |
- |
hypothetical protein POPTR_0015s12340g [Populus trichocarpa] |
| 18 |
Hb_022815_020 |
0.1744536776 |
- |
- |
PREDICTED: uncharacterized protein LOC105650107 [Jatropha curcas] |
| 19 |
Hb_001172_030 |
0.1752709869 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 7.2-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000567_300 |
0.1762973283 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |