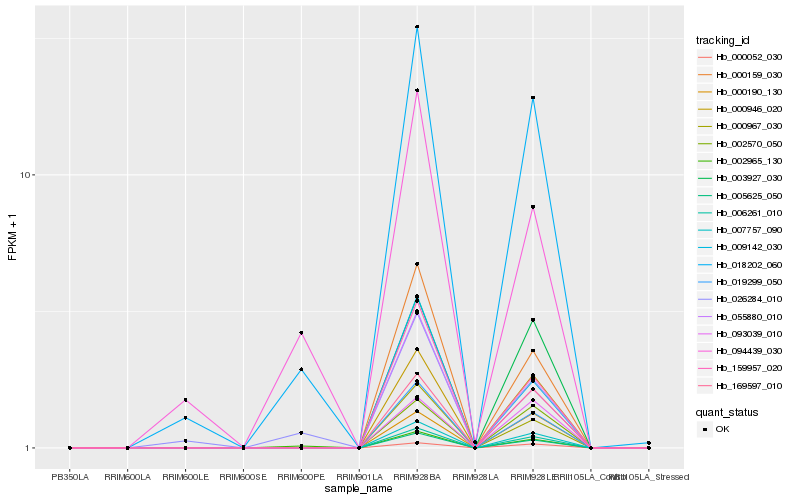
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018202_060 |
0.0 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Jatropha curcas] |
| 2 |
Hb_007757_090 |
0.1097373167 |
- |
- |
PREDICTED: uncharacterized protein LOC105793478 [Gossypium raimondii] |
| 3 |
Hb_006261_010 |
0.109882886 |
- |
- |
PREDICTED: uncharacterized protein LOC104110884 [Nicotiana tomentosiformis] |
| 4 |
Hb_005625_050 |
0.110058493 |
- |
- |
hypothetical protein PRUPE_ppa024499mg, partial [Prunus persica] |
| 5 |
Hb_019299_050 |
0.1137961588 |
- |
- |
- |
| 6 |
Hb_026284_010 |
0.1153249442 |
- |
- |
- |
| 7 |
Hb_000946_020 |
0.1169343481 |
- |
- |
hypothetical protein PRUPE_ppa023262mg [Prunus persica] |
| 8 |
Hb_002965_130 |
0.1208583867 |
- |
- |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 9 |
Hb_169597_010 |
0.1276816366 |
- |
- |
PREDICTED: uncharacterized protein LOC104903465 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_000967_030 |
0.1300104035 |
- |
- |
- |
| 11 |
Hb_003927_030 |
0.1303988278 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_000052_030 |
0.131121323 |
- |
- |
- |
| 13 |
Hb_000159_030 |
0.1401843027 |
- |
- |
putative glycosyltransferase, partial [Aralia elata] |
| 14 |
Hb_094439_030 |
0.1410379722 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g38780 [Eucalyptus grandis] |
| 15 |
Hb_159957_020 |
0.1412440378 |
- |
- |
- |
| 16 |
Hb_002570_050 |
0.1450552263 |
- |
- |
PREDICTED: uncharacterized protein LOC104897677 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_055880_010 |
0.1568687403 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_093039_010 |
0.1575474625 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 19 |
Hb_009142_030 |
0.1590048439 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000190_130 |
0.1601683306 |
- |
- |
cysteine protease inhibitor [Populus tremula] |