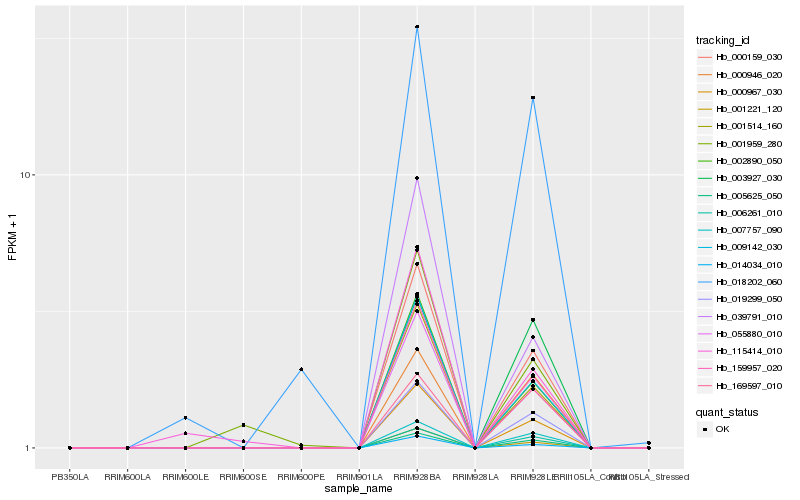
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_159957_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000159_030 |
0.0017093681 |
- |
- |
putative glycosyltransferase, partial [Aralia elata] |
| 3 |
Hb_000967_030 |
0.0193773022 |
- |
- |
- |
| 4 |
Hb_055880_010 |
0.0234234548 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_009142_030 |
0.0264300552 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_001514_160 |
0.0283740104 |
- |
- |
hypothetical protein JCGZ_16070 [Jatropha curcas] |
| 7 |
Hb_014034_010 |
0.0330721577 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 8 |
Hb_002890_050 |
0.0340427888 |
- |
- |
- |
| 9 |
Hb_001221_120 |
0.0365035008 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_019299_050 |
0.0592174171 |
- |
- |
- |
| 11 |
Hb_005625_050 |
0.0810003556 |
- |
- |
hypothetical protein PRUPE_ppa024499mg, partial [Prunus persica] |
| 12 |
Hb_007757_090 |
0.0887712824 |
- |
- |
PREDICTED: uncharacterized protein LOC105793478 [Gossypium raimondii] |
| 13 |
Hb_006261_010 |
0.095160517 |
- |
- |
PREDICTED: uncharacterized protein LOC104110884 [Nicotiana tomentosiformis] |
| 14 |
Hb_039791_010 |
0.1088129003 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000946_020 |
0.1298912857 |
- |
- |
hypothetical protein PRUPE_ppa023262mg [Prunus persica] |
| 16 |
Hb_018202_060 |
0.1412440378 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Jatropha curcas] |
| 17 |
Hb_115414_010 |
0.1531675232 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 18 |
Hb_001959_280 |
0.1533936891 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 19 |
Hb_169597_010 |
0.1547437591 |
- |
- |
PREDICTED: uncharacterized protein LOC104903465 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_003927_030 |
0.1599005778 |
- |
- |
unnamed protein product [Coffea canephora] |