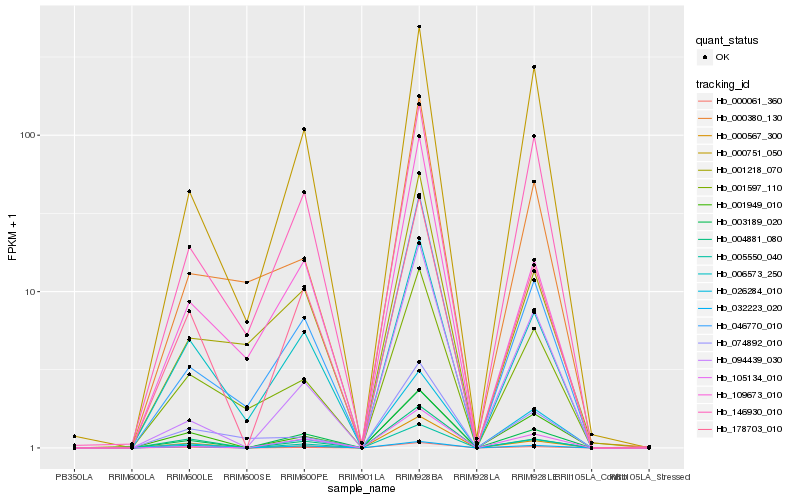
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005550_040 |
0.0 |
- |
- |
PREDICTED: epoxide hydrolase 4-like [Jatropha curcas] |
| 2 |
Hb_003189_020 |
0.0608072579 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 3 |
Hb_178703_010 |
0.0866859256 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 4 |
Hb_000061_360 |
0.0867687204 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 5 |
Hb_001949_010 |
0.0956053468 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 6 |
Hb_046770_010 |
0.1006889169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_105134_010 |
0.1017631336 |
- |
- |
hypothetical protein JCGZ_16347 [Jatropha curcas] |
| 8 |
Hb_000751_050 |
0.1198866825 |
- |
- |
PREDICTED: extensin, partial [Solanum lycopersicum] |
| 9 |
Hb_094439_030 |
0.1218968934 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g38780 [Eucalyptus grandis] |
| 10 |
Hb_006573_250 |
0.1223579981 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 11 |
Hb_004881_080 |
0.127072117 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 1 [Populus euphratica] |
| 12 |
Hb_026284_010 |
0.1304337797 |
- |
- |
- |
| 13 |
Hb_032223_020 |
0.1318097248 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_000567_300 |
0.1326419651 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 15 |
Hb_001597_110 |
0.1410015577 |
- |
- |
- |
| 16 |
Hb_109673_010 |
0.1514285401 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 17 |
Hb_146930_010 |
0.1542760093 |
- |
- |
- |
| 18 |
Hb_000380_130 |
0.1565551185 |
- |
- |
Phenylalanine ammonia-lyase, putative [Ricinus communis] |
| 19 |
Hb_001218_070 |
0.1632239071 |
- |
- |
PREDICTED: sugar transport protein 5-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_074892_010 |
0.1642163817 |
- |
- |
Flavonol 3-sulfotransferase, putative [Ricinus communis] |