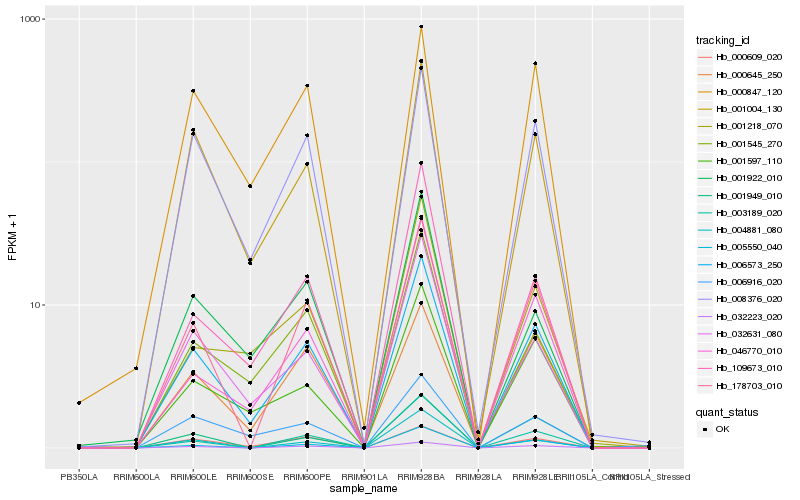
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006573_250 |
0.0 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 2 |
Hb_178703_010 |
0.0901600339 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 3 |
Hb_001004_130 |
0.091450106 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 4 |
Hb_001597_110 |
0.0988394793 |
- |
- |
- |
| 5 |
Hb_032223_020 |
0.1000839118 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_008376_020 |
0.1032433168 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 7 |
Hb_032631_080 |
0.1139387388 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_001545_270 |
0.114723587 |
- |
- |
laccase, putative [Ricinus communis] |
| 9 |
Hb_005550_040 |
0.1223579981 |
- |
- |
PREDICTED: epoxide hydrolase 4-like [Jatropha curcas] |
| 10 |
Hb_003189_020 |
0.1238190098 |
- |
- |
PREDICTED: probable glutathione S-transferase [Jatropha curcas] |
| 11 |
Hb_046770_010 |
0.129394372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001922_010 |
0.1308600639 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 13 |
Hb_000609_020 |
0.1336358425 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 14 |
Hb_109673_010 |
0.1339789962 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 15 |
Hb_001218_070 |
0.134329766 |
- |
- |
PREDICTED: sugar transport protein 5-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001949_010 |
0.1345118595 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 17 |
Hb_000645_250 |
0.1352207568 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 18 |
Hb_004881_080 |
0.1364358556 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 1 [Populus euphratica] |
| 19 |
Hb_000847_120 |
0.1391549308 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 20 |
Hb_006916_020 |
0.1395461409 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |