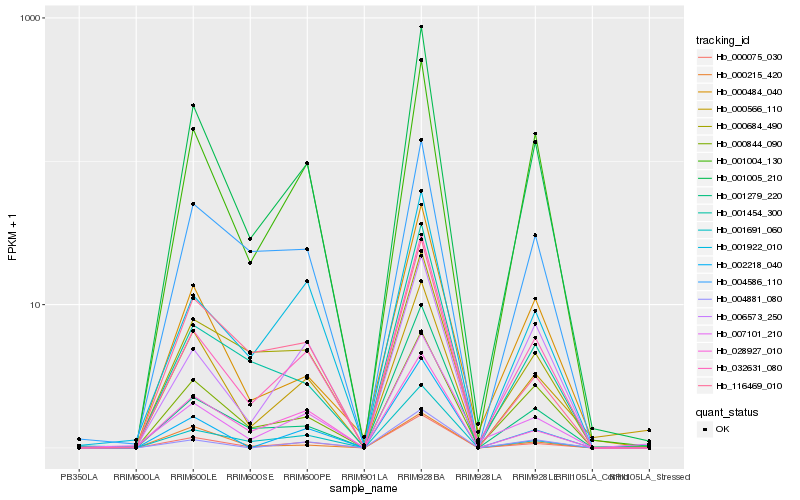
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001005_210 |
0.0 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 2 |
Hb_000075_030 |
0.0590733124 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C10-like [Fragaria vesca subsp. vesca] |
| 3 |
Hb_032631_080 |
0.0713460098 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_001004_130 |
0.1054245583 |
- |
- |
PREDICTED: chalcone synthase 3 [Jatropha curcas] |
| 5 |
Hb_000484_040 |
0.1081689918 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000844_090 |
0.1123117721 |
- |
- |
PREDICTED: cytochrome P450 82C4-like [Jatropha curcas] |
| 7 |
Hb_001691_060 |
0.1141102627 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 8 |
Hb_000566_110 |
0.1203085283 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Prunus mume] |
| 9 |
Hb_002218_040 |
0.1213828957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001279_220 |
0.1300499769 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_004881_080 |
0.1318026179 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 1 [Populus euphratica] |
| 12 |
Hb_001922_010 |
0.1328119814 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 13 |
Hb_001454_300 |
0.1329344319 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 14 |
Hb_007101_210 |
0.1342547206 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000215_420 |
0.1393385589 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 16 |
Hb_028927_010 |
0.1403024285 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 17 |
Hb_004586_110 |
0.1438323582 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 18 |
Hb_000684_490 |
0.1466587353 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 44 [Jatropha curcas] |
| 19 |
Hb_006573_250 |
0.1474651724 |
- |
- |
PREDICTED: ABC transporter G family member 8 [Jatropha curcas] |
| 20 |
Hb_116469_010 |
0.1514334987 |
- |
- |
PREDICTED: uncharacterized protein LOC105647834 [Jatropha curcas] |