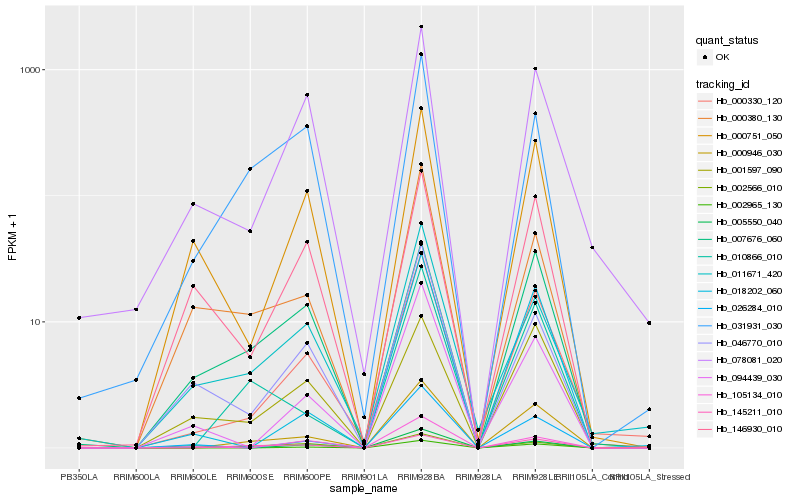
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000330_120 |
0.0 |
- |
- |
PREDICTED: bifunctional dihydroflavonol 4-reductase/flavanone 4-reductase-like [Nelumbo nucifera] |
| 2 |
Hb_000946_030 |
0.1136473894 |
- |
- |
hypothetical protein JCGZ_10707 [Jatropha curcas] |
| 3 |
Hb_002965_130 |
0.1249834101 |
- |
- |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 4 |
Hb_078081_020 |
0.134349336 |
- |
- |
- |
| 5 |
Hb_094439_030 |
0.14176743 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g38780 [Eucalyptus grandis] |
| 6 |
Hb_026284_010 |
0.1485226477 |
- |
- |
- |
| 7 |
Hb_000751_050 |
0.1527046036 |
- |
- |
PREDICTED: extensin, partial [Solanum lycopersicum] |
| 8 |
Hb_011671_420 |
0.1597669094 |
- |
- |
hypothetical protein POPTR_0015s12340g [Populus trichocarpa] |
| 9 |
Hb_105134_010 |
0.1638303171 |
- |
- |
hypothetical protein JCGZ_16347 [Jatropha curcas] |
| 10 |
Hb_046770_010 |
0.165258101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_018202_060 |
0.1682106478 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Jatropha curcas] |
| 12 |
Hb_001597_090 |
0.170381702 |
- |
- |
- |
| 13 |
Hb_146930_010 |
0.178403965 |
- |
- |
- |
| 14 |
Hb_145211_010 |
0.1812822633 |
- |
- |
hypothetical protein M569_05648, partial [Genlisea aurea] |
| 15 |
Hb_010866_010 |
0.1833449154 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Vitis vinifera] |
| 16 |
Hb_000380_130 |
0.1843634537 |
- |
- |
Phenylalanine ammonia-lyase, putative [Ricinus communis] |
| 17 |
Hb_002566_010 |
0.1852758553 |
- |
- |
PREDICTED: uncharacterized protein LOC105643984 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007676_060 |
0.1905278442 |
- |
- |
PREDICTED: cellulose synthase-like protein E2 [Pyrus x bretschneideri] |
| 19 |
Hb_031931_030 |
0.1935170544 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 20 |
Hb_005550_040 |
0.1965033306 |
- |
- |
PREDICTED: epoxide hydrolase 4-like [Jatropha curcas] |