| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
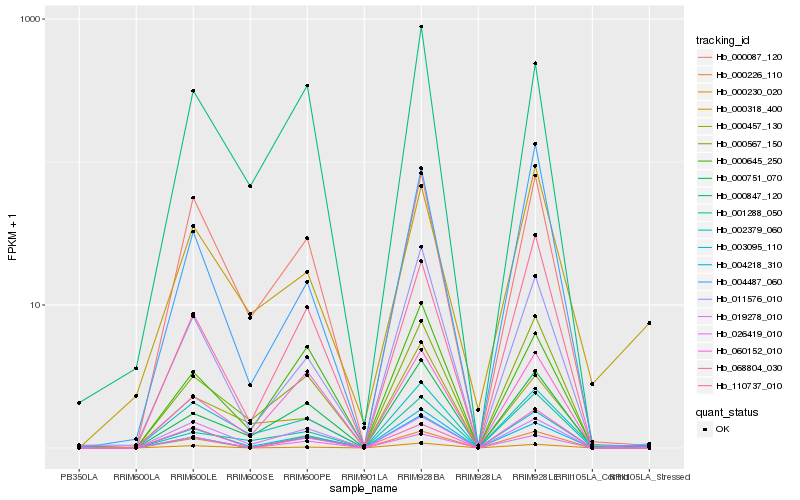
Hb_003095_110 |
0.0 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 2 |
Hb_004218_310 |
0.1256807876 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Camelina sativa] |
| 3 |
Hb_011576_010 |
0.1339127197 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 4 |
Hb_000567_150 |
0.139799205 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 5 |
Hb_000087_120 |
0.1399565341 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 6 |
Hb_000230_020 |
0.1418522608 |
- |
- |
cysteine synthase, putative [Ricinus communis] |
| 7 |
Hb_000318_400 |
0.1464440124 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000751_070 |
0.1508791312 |
- |
- |
- |
| 9 |
Hb_110737_010 |
0.1547397518 |
- |
- |
PREDICTED: aldose 1-epimerase-like [Jatropha curcas] |
| 10 |
Hb_002379_060 |
0.1590591711 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 11 |
Hb_068804_030 |
0.1620794736 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 12 |
Hb_001288_050 |
0.1652861235 |
- |
- |
- |
| 13 |
Hb_004487_060 |
0.1686550514 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_026419_010 |
0.172117574 |
- |
- |
PREDICTED: cyclin-D3-1-like [Jatropha curcas] |
| 15 |
Hb_000645_250 |
0.1778631711 |
- |
- |
PREDICTED: putative beta-D-xylosidase [Jatropha curcas] |
| 16 |
Hb_000847_120 |
0.1783824794 |
- |
- |
hypothetical protein JCGZ_15115 [Jatropha curcas] |
| 17 |
Hb_060152_010 |
0.1785405342 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 18 |
Hb_019278_010 |
0.1790211962 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 19 |
Hb_000226_110 |
0.1848747473 |
- |
- |
- |
| 20 |
Hb_000457_130 |
0.1855304481 |
- |
- |
hypothetical protein AALP_AA3G009100 [Arabis alpina] |