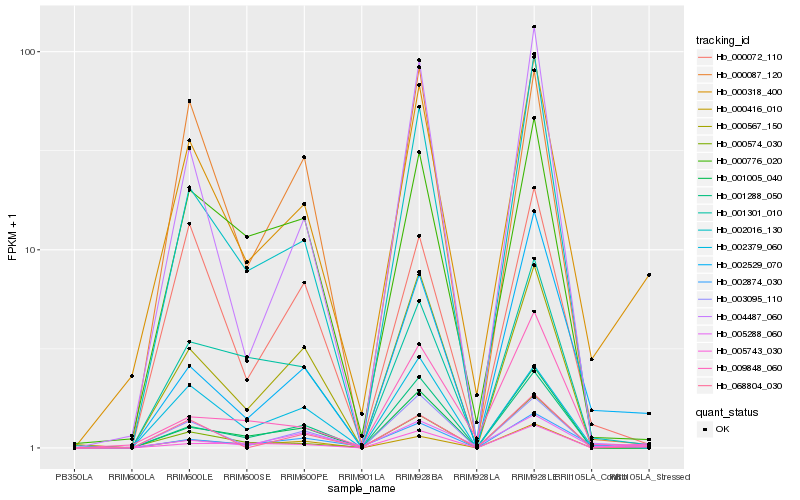
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_400 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003095_110 |
0.1464440124 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 3 |
Hb_002016_130 |
0.1728037892 |
desease resistance |
Gene Name: ABC_membrane |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 4 |
Hb_000776_020 |
0.173550696 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_25594 [Jatropha curcas] |
| 5 |
Hb_000574_030 |
0.1769630018 |
desease resistance |
Gene Name: Glyco_hydro_38 |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 6 |
Hb_005288_060 |
0.1821580924 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 7 |
Hb_000567_150 |
0.1823645585 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 8 |
Hb_000072_110 |
0.1824185242 |
- |
- |
PREDICTED: U-box domain-containing protein 35 [Jatropha curcas] |
| 9 |
Hb_005743_030 |
0.183004678 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 10 |
Hb_001005_040 |
0.1852846751 |
- |
- |
- |
| 11 |
Hb_002379_060 |
0.1856926198 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 12 |
Hb_000087_120 |
0.1871608608 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 13 |
Hb_004487_060 |
0.1873693024 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_001301_010 |
0.1882737905 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 15 |
Hb_001288_050 |
0.1885584181 |
- |
- |
- |
| 16 |
Hb_009848_060 |
0.1898755896 |
- |
- |
unknown [Glycine max] |
| 17 |
Hb_002529_070 |
0.1903949348 |
- |
- |
PREDICTED: putative late blight resistance protein homolog R1B-19 [Pyrus x bretschneideri] |
| 18 |
Hb_068804_030 |
0.1906991227 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 19 |
Hb_002874_030 |
0.1928646755 |
- |
- |
hypothetical protein JCGZ_07001 [Jatropha curcas] |
| 20 |
Hb_000416_010 |
0.1929974901 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |